Anti-PD-L1 nano antibody and application thereof
A PD-L1 and nanobody technology, applied in the field of biomedicine and molecular biology, can solve the problems of harsh antigen requirements, singleness, long immunization time, etc., and achieve the effect of excellent stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1: the design of novel V-NAR frame
[0035]Access 2YWZ_A, AAP86762, 4HGK_C, AAN75852, AAM33845, Lep-12E1, ABY64741, Tom70, etc. through PDB (https: / / www.rcsb.org) and NCBI (https: / / www.ncbi.nlm.nih.gov) databases V-NAR Nanobody amino acid sequence, and the V-NAR amino acid sequence was analyzed using clustalw (https: / / www.ebi.ac.uk) and WebLogo (http: / / weblogo.berkeley.edu / logo.cgi). The amino acid sequences of the four FR regions of the V-NAR framework were determined according to the frequency of amino acids at the corresponding positions in the framework region on the sequence alignment results. For the amino acid sequences of the CDR1, HV2 and HV4 regions of the V-NAR framework, not only the sequence comparison results, but also the amino acids at specific sites in the V-NAR that are conducive to antibody stability should be referred to, and finally the CDR1 of the V-NAR framework should be determined. , the amino acid sequences of the HV2 and HV4 region...
Embodiment 2
[0036] Example 2: Construction and evaluation of V-NAR phage library
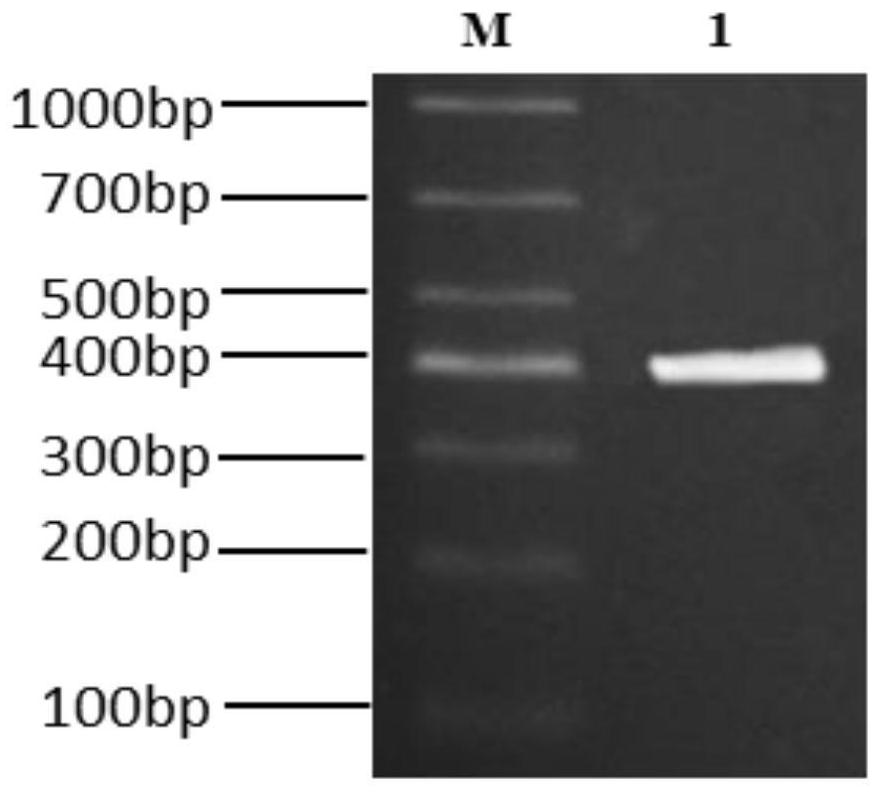
[0037] The full-length gene fragment of V-NAR was amplified by overlap extension PCR, a total of 3 rounds of PCR. The first round of PCR amplification obtained DNA fragments in the FR1-FR3 region. The DNA fragment of the CDR3-FR4 region was obtained in the second round of PCR amplification, and the complete V-NAR full antibody gene fragment was obtained in the third round of PCR amplification ( figure 2 ). Subsequently, it and the pCANTAB5E phagemid vector were digested with Sfi I and Not I endonucleases, ligated and electrotransformed into Escherichia coli TG1. Then use 2×TY medium to dilute the V-NAR synthetic library according to 10 -1 、10 -2 、10 -3 to 10 -8 (10-fold) gradient dilution, take 100 μL of each concentration of bacterial solution and spread it on a 2×TY plate containing Amp, and detect the storage capacity of V-NAR according to the dilution factor and the number of single colonies on t...
Embodiment 3
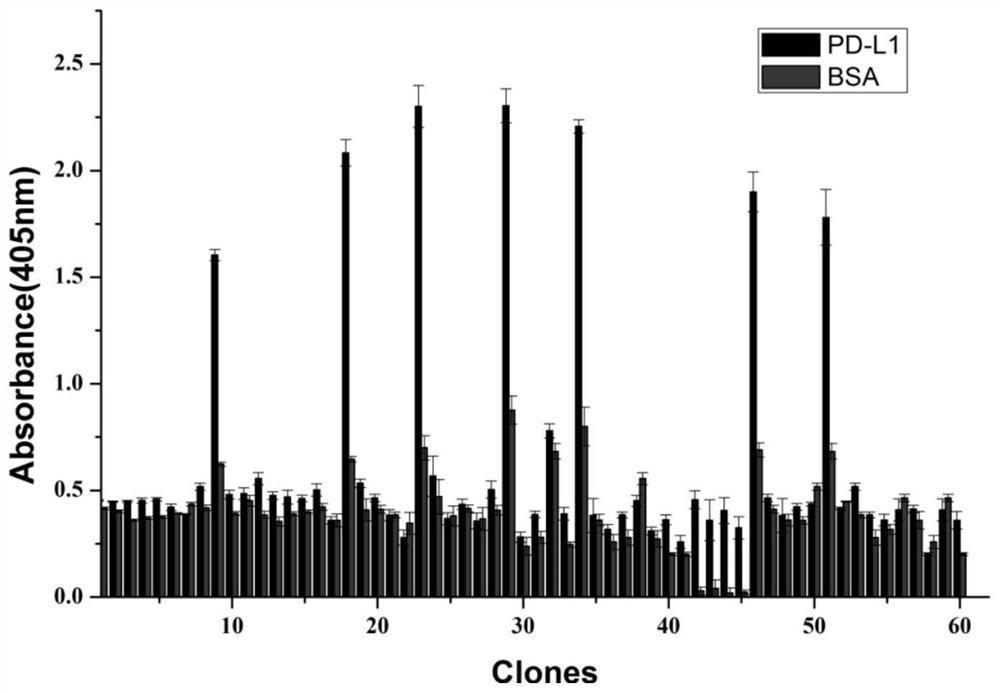
[0040] Example 3: Screening and preliminary identification of PD-L1-specific nanobodies
[0041] Dilute PD-L1 protein and BSA with NaHCO 3 The buffer was diluted to a concentration of 100 μg / mL, PD-L1 protein was used as the experimental group, and BSA was used as the control group. Add 150 μL to each well, set up 3 replicate wells, and incubate overnight at 4°C. Then the reaction wells were washed with TBST (0.1%) buffer and blocked with 3% skimmed milk at 4° C. for 2 h. After washing, 100 μL of phage library solution was added and incubated at room temperature for 60 min. Then, after washing 10 times with TBST buffer, 200 μL of eluate was added to the wells, and incubated at room temperature for 10 min. After the incubation, 15 μL of neutralization buffer was added to each well, which was the positive phage obtained in the first round of screening, and the titer was determined. Dilute it with 2×TY medium, infect the logarithmic phase TG1 glycerolbacterium, and incubate f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com