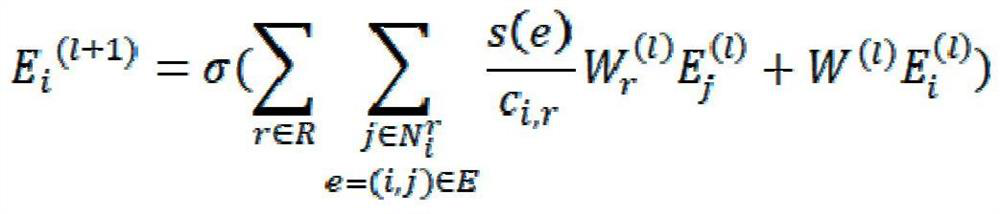New coronavirus target prediction and drug discovery method based on graph representation learning
A technology of target prediction and graph representation, applied in the field of bioinformatics to achieve the effect of speeding up drug research and development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] The present invention will be further described below in conjunction with the accompanying drawings and embodiments.
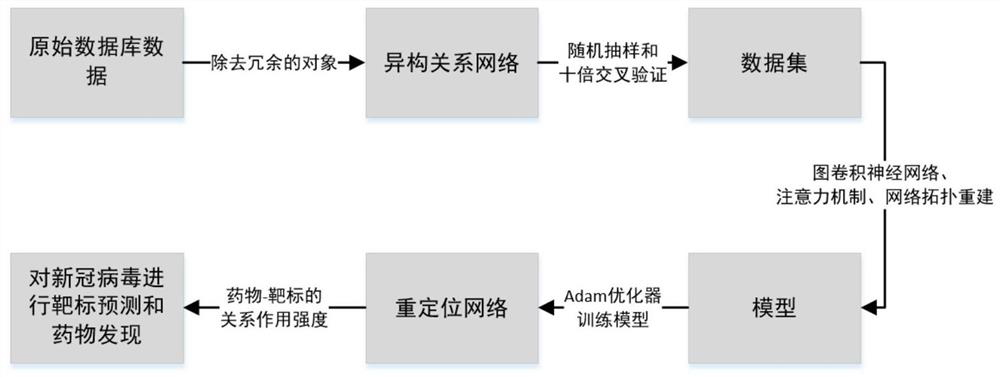
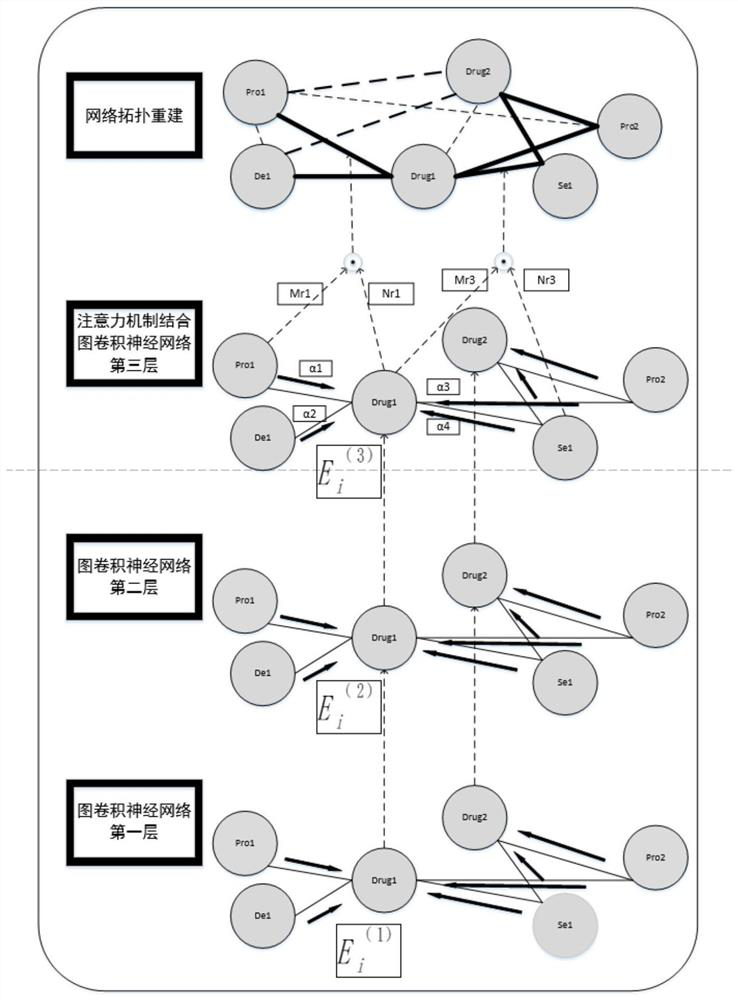
[0035] Such as figure 1 As shown, the steps of the method are specifically described below in conjunction with an example:
[0036]The first step is to prepare a heterogeneous network data set; construct a heterogeneous relationship network including the relationship between drugs, targets, side effects, and diseases, where the drug-target interaction and drug-drug interaction network are based on the DRUGBANK database, and the target- The target interaction network is based on the HPRD database, the drug-disease association and target-disease association network is based on the CTD database, and the drug-side effect association network is based on the SIDER database; in the process of constructing heterogeneous network datasets, the common object. For example, if there is drug x in the drug-drug interaction network, but there is no drug x in the drug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com