Method for quickly and quantitatively detecting target DNA
A quantitative detection and target technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of many operation steps, influence of target DNA, long time consumption, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0071] Cell fusion and preparation of primary antibody: Aseptically prepare mouse splenocyte suspension, mix with mouse myeloma cell SP2 / 0 at a ratio of 10:1, fuse with PEG, screen with HAT medium for 7 days, then replace with HT Culture medium, when there are obvious cell clusters in the wells, screen with the method consistent with the detection of serum titer, and the screened positive wells are subjected to 3 times of limited dilution to prepare monoclonal cell lines and expand them After culture, mouse ascites was prepared and purified with ProteinA to obtain the primary antibody.
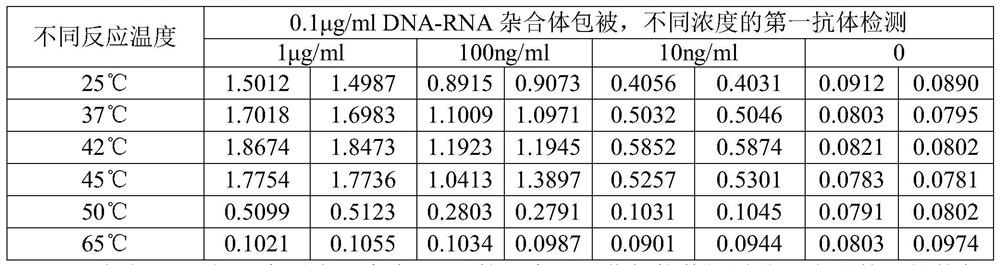
[0072] Validation of the primary antibody:
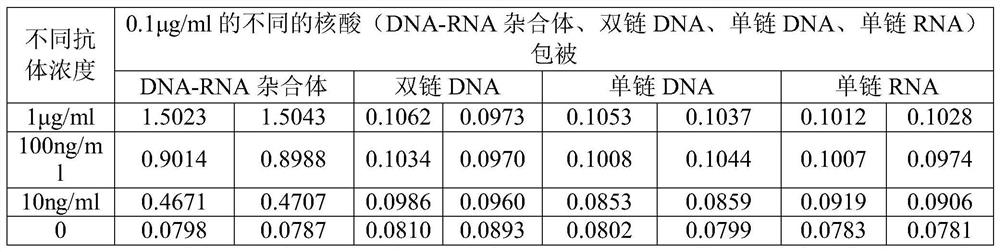
[0073]Verification method 1: Use 0.1 μg / ml of different nucleic acids (DNA-RNA hybrids, double-stranded DNA, single-stranded DNA, single-stranded RNA) to duplicate 2 wells, coat at 4°C overnight, and block with gelatin at 37°C for 2 hours , react with different antibody concentrations (1μg / ml, 100ng / ml, 10ng / ml, 0) for 1 hour at room temperature (2...
Embodiment 1
[0103] Embodiment 1: A method for rapid quantitative detection of target DNA, comprising the steps of:
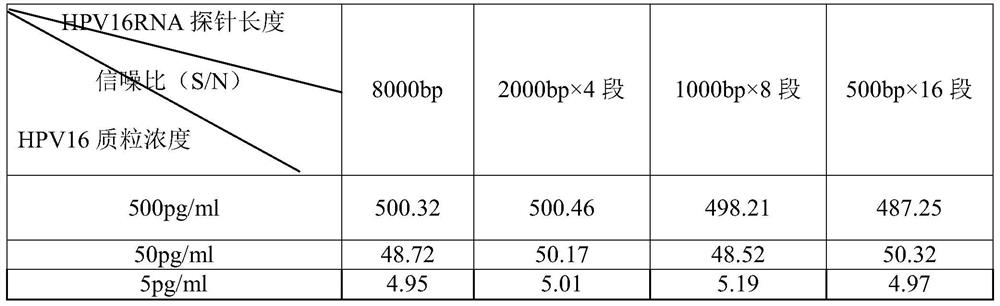
[0104] Step 1, denaturation: react the sodium hydroxide solution with the cervical exfoliated cell sample at 65°C for 30 minutes, and the concentration of the sodium hydroxide solution is 1.75mol / L, and the volume ratio of the sodium hydroxide solution to the sample to be tested is 1:2, crush the cells of the sample to be tested, denature the protein, degrade the RNA, and decompose the DNA into single-stranded DNA to obtain a denatured sample; step 2, hybridization: the denatured sample obtained in step 1 Mix with the HPV16 RNA probe (full-length single-stranded nucleic acid probe for target DNA, 8000bp in length) stored in the nucleic acid preservation solution with a pH value of 3.5-4.0, the volume of the HPV16 RNA probe and the denatured sample The ratio is 1:3, hybridize for 45 minutes under the condition of pH value 7.0-7.4 and temperature 65°C, so that the HPV16 RNA p...
Embodiment 2
[0112] Embodiment 2: A method for rapid quantitative detection of target DNA, the difference from Example 1 is that the HPV16 RNA probe consists of 4 sections of full-length discontinuous HPV16 RNA probes, and the full-length discontinuous HPV16 RNA probes The length is 2000bp.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com