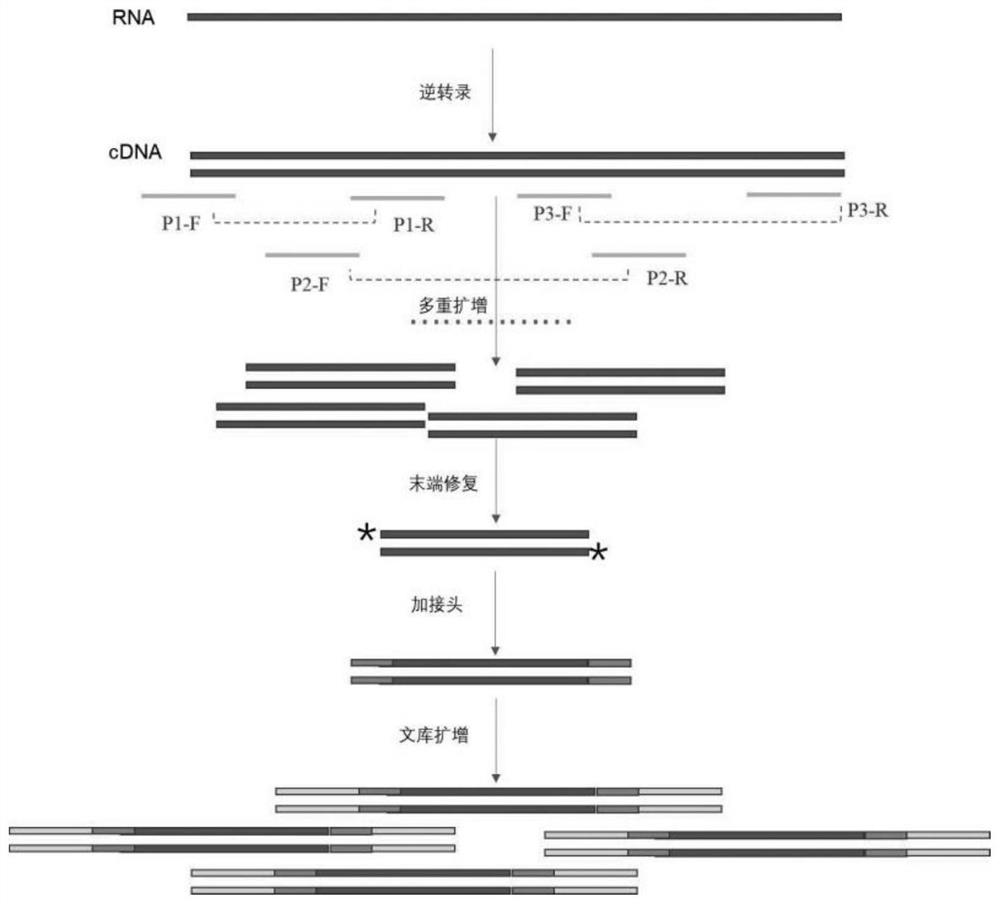
Method for constructing library for full-length genome sequencing of novel coronavirus SARS-CoV-2
A genome sequencing, coronavirus technology, applied in chemical libraries, biochemical equipment and methods, combinatorial chemistry, etc., can solve the problems of inability to detect and analyze virus mutation, incomplete gene region, poor specificity, etc., to improve connection efficiency, The effect of high sensitivity and high detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0051] The present invention will be described in detail below in conjunction with specific embodiments.
[0052] The present invention provides a method for constructing a library for sequencing the full-length genome of novel coronavirus SARS-CoV-2, which specifically includes the following steps:
[0053] S1. cDNA synthesis: extract viral RNA, random primer hexamer, viral RNA, and nuclease-free water to prepare a reaction system, mix them in a PCR tube on ice, and place them in a PCR instrument for the first reaction. After the reaction, the PCR tube Take it out, add Reverse Transcription mix G2 (reverse transcriptase reaction solution), mix well and put it in the PCR instrument again to carry out the reverse transcription product from the secondary reaction according to the set temperature program. The specific operation method is as follows:
[0054] (1) Take 1pg ~ 500ng of the extracted viral RNA and configure the system on ice in a clean PCR tube according to the compos...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com