Diagnostic marker and application thereof in COVID-19 diagnosis and coronavirus previous infection detection
A COVID-19, COVID19-V001 technology, applied in the field of biomedicine, can solve the problems of difficult expression of protein structure, difficulty in stable storage, and increased difficulty in protein purification, so as to improve the effect of diagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
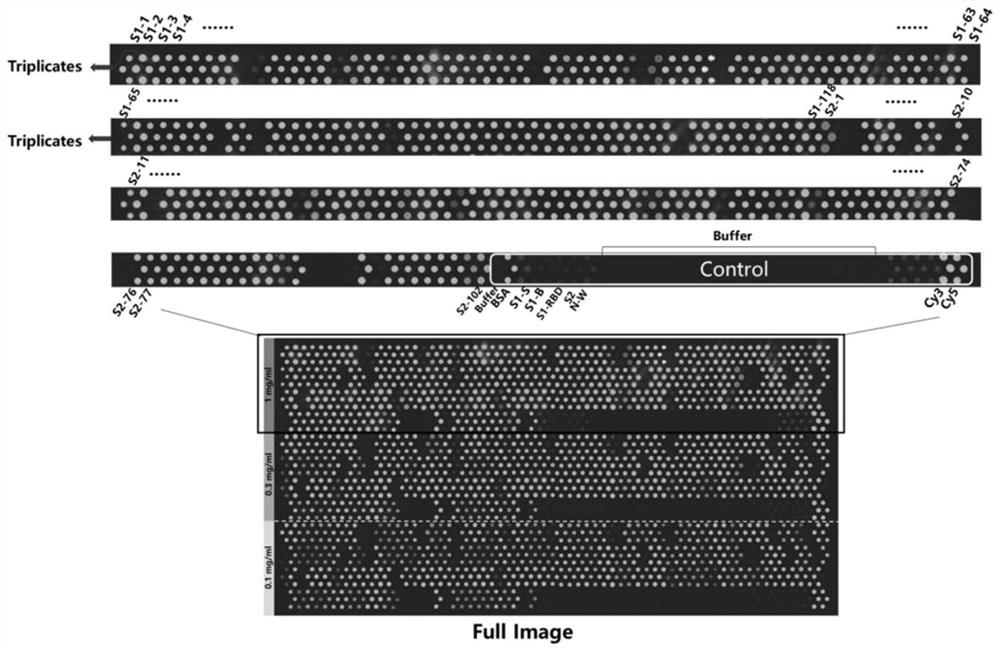
[0070] Example 1 Detection of serum from recovered patients with COVID-19 in the form of a small peptide chip
[0071] 1. Peptide processing and coupling
[0072] 1.1 Preparation of samples: According to the condition that each peptide is 12aa in length and there is an overlapping length of 6aa between every two peptides, there are 220 peptides in S protein. Among them, the S1 region contains 118 peptides, and the S2 region contains 102 peptides, including the peptide segment FKEELDKYFKNH in the present invention, which was finally synthesized and purified by Jill Biochemical (Shanghai) Co., Ltd., and a Cys couple was added to the N-terminus of each purified peptide. Linked to BSA, 197 successful co-couplings (coupling products).
[0073] The specific coupling steps are as follows:
[0074] ① Dissolve 10mg of BSA in 1mL of PBS Buffer at a concentration of 10mg / mL.
[0075] ② Dissolve 10 μL SMCC (weigh 1 mg SMCC and dissolve in 10 μL DMSO) in the BSA solution, and place it...
Embodiment 2
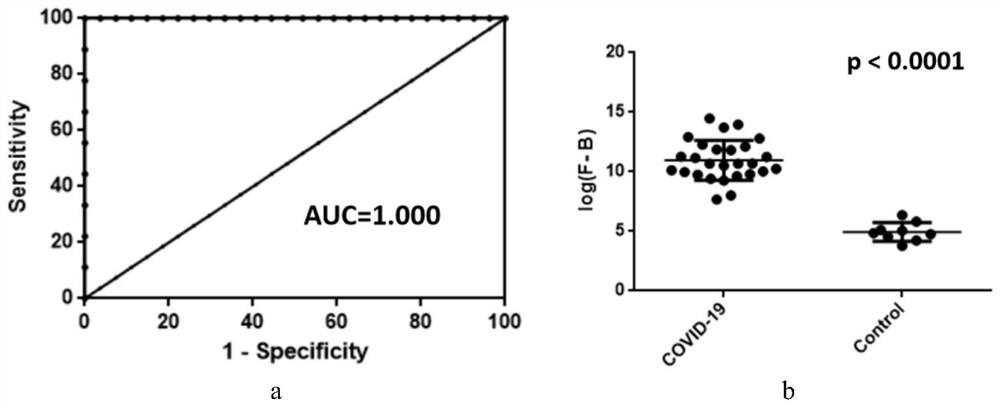
[0148] Example 2 Verification of a large number of serum samples by small peptide chips
[0149] 1. Peptide processing and coupling
[0150] This step is the same as in Example 1
[0151] 2. Incubation of chip and serum
[0152] 2.1 Preparation of required reagents
[0153] Blocking solution: 3g BSA, add 100mL 1x PBS solution (diluted from 10x PBS), mix well.
[0154] Incubation solution: 1x PBST solution (0.1% Tween20).
[0155] Washing solution: 1x PBST.
[0156] The 10x PBS (1L) formulation was prepared using the components shown in Table 1 in Example 1.
[0157] 2.2 Serum experiment
[0158] a. Block the chip: Prepare 30 mL of blocking solution (3% BSA in PBSbuffer) in a chip box that can hold 4 chips. Take the chip prepared in step 1 from -80°C to 4°C and rewarm at room temperature. After the chip enters the blocking solution, quickly shake the chip in parallel and reverse it in the blocking solution. Place the sealing box on a side swing shaker for 20- At 30rpm...
Embodiment 3
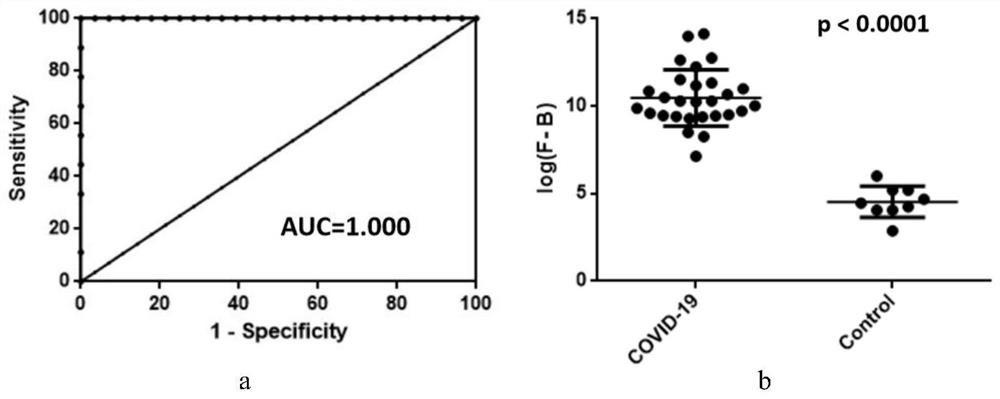
[0199] Example 3 By performing site mutation on the candidate peptide COVID19-V001
[0200] 1. Peptide processing and coupling
[0201] The last 7 amino acids (DKYFKNH) of the candidate peptide COVID19-V001 obtained in Examples 1 and 2 were mutated into alanine (A) one by one to generate 7 mutant types (the last 7 amino acid sequences after mutation were AKYFKNH, AKYFKNH, DAYFKNH, DKAFKNH, DKYAKNH, DKYFANH, DKYFKAH, DKYFKNA), 7 mutant types and 1 wild type were coupled to BSA (the coupling method was the same as in Example 1).
[0202] 2. Preparation of the chip: the method is the same as that of the chip in Example 1
[0203] 3. Incubation of chip and serum
[0204] 3.1 Preparation of required reagents
[0205] Blocking solution: 3g BSA, add 100mL 1x PBS solution (diluted from 10x PBS), mix well.
[0206] Incubation solution: 1x PBST solution (0.1% Tween20).
[0207] Washing solution: 1x PBST.
[0208] The 10x PBS (1L) formulation was prepared using the components sho...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com