Primary hepatocellular carcinoma molecular typing and survival risk gene group, diagnosis product and application
A hepatocellular carcinoma and molecular typing technology, which is applied in the field of primary hepatocellular carcinoma molecular typing, survival risk gene group and diagnostic products and applications, can solve the problems that clinicians and patients cannot guide treatment and predict prognosis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
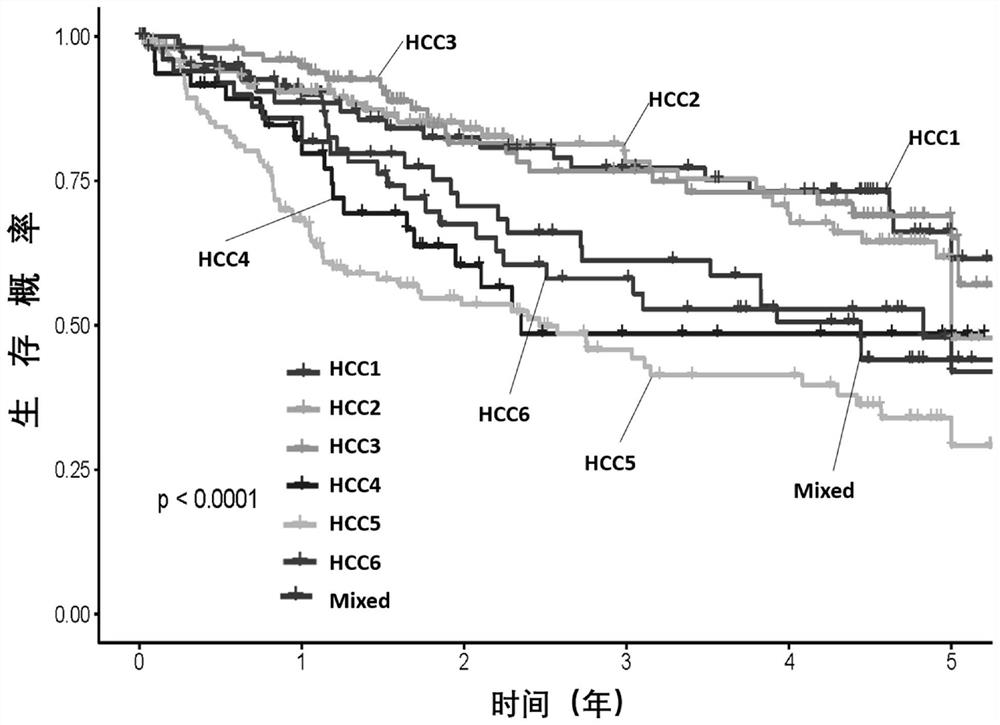
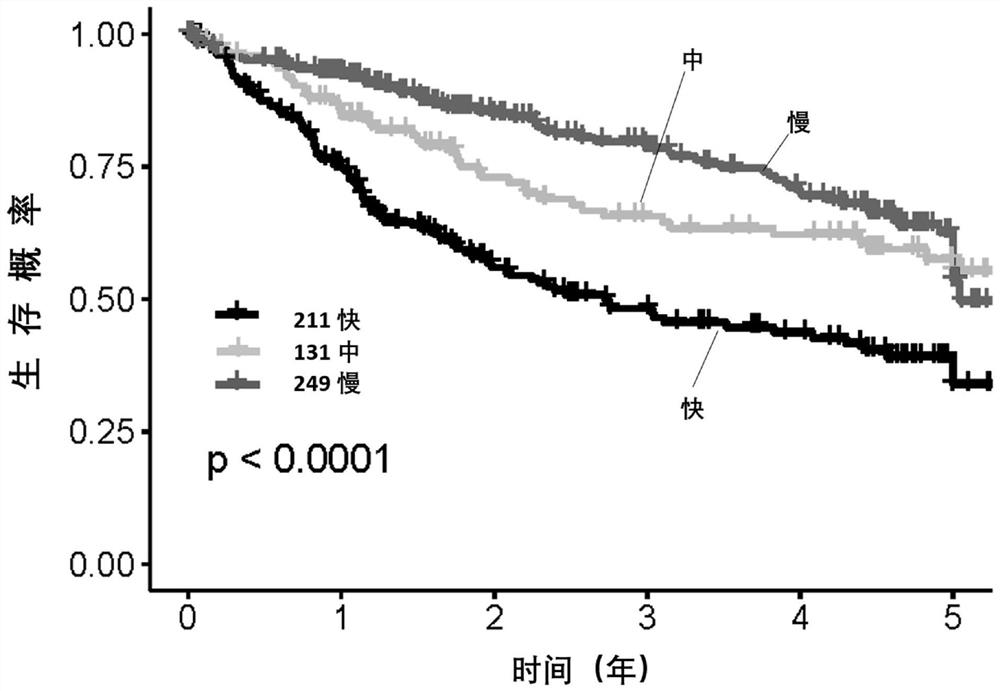
[0115] Evaluation of subtypes of liver cancer and screening of survival risk-related gene groups
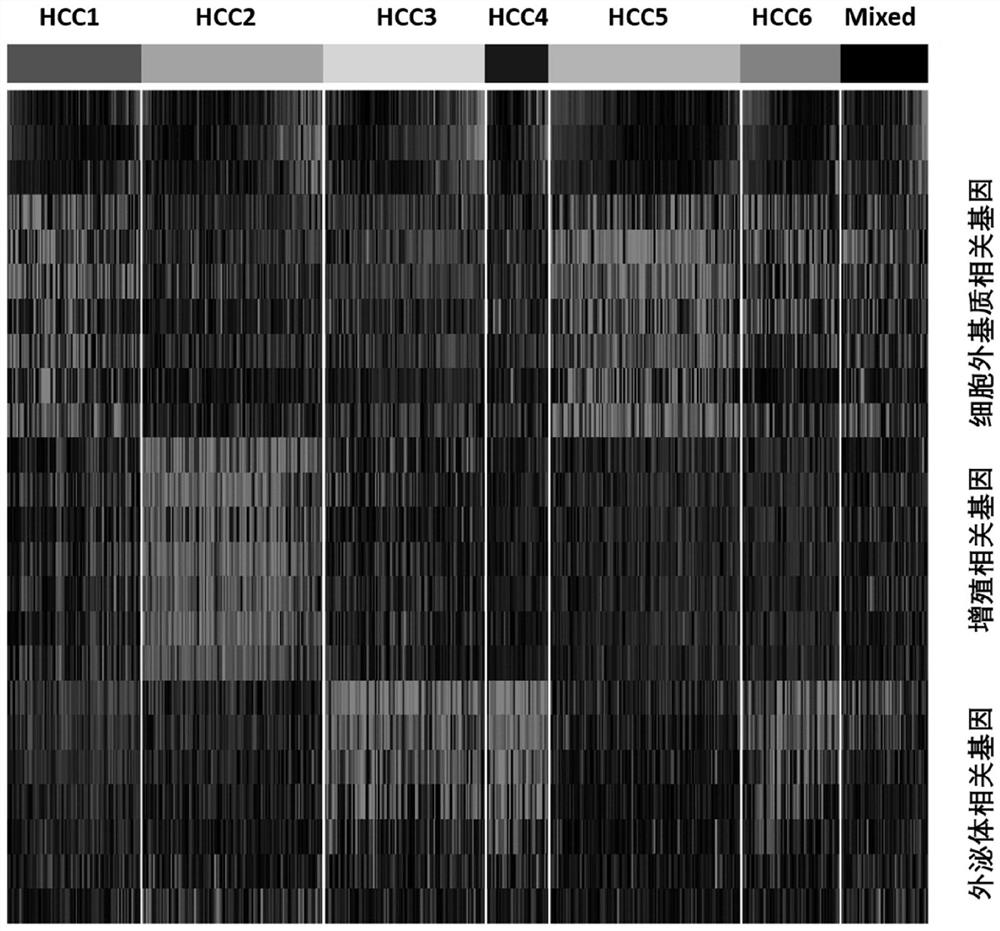
[0116] Methods: 370 cases in the TCGA database with complete Based on the expression level of primary liver cancer genes in clinical information, cell proliferation-related genes, immune-related genes, intercellular matrix-related genes, exosome-related genes, and FTH family genes that are closely related to the survival risk of liver cancer were screened out, and each group of genes Calculate and optimize the genes with a large contribution rate to typing and survival risk.
[0117] Results: A total of 62 genes and 6 reference genes related to liver cancer subtype and survival risk were screened, that is, 68 gene test combinations. See Table 1 for the list of genes.
[0118] The validity and stability of the 68 genes screened were verified in the Affymatrics gene chip expression profile data of 221 cases of primary liver cancer.
[0119] Table 1: Molecular classification of H...
Embodiment 2
[0124] Molecular typing and survival risk assessment of liver cancer by next-generation sequencing
[0125] Experimental method: A 68-gene test combination was used, of which 62 liver cancer subtypes and survival risk-related gene groups (proliferation-related genes BIRC5, BUB1, CCNB2, CDC20, CDC6, CDK1, E2F1, FOXM1, GINS1, KIF18B, KIF2C, MKI67 , MYBL2, PLK1, TOP2A, TPX2, TROAP, UBE2C, immune-related genes CD2, CD74, CD8A, PLA2G2D, SLAMF7, extracellular matrix-related genes ABCB11, ADH1B, ALDOB, APOF, AQP9, BHMT, CYP2A6, F9, FETUB, GLYAT, GYS2, HP, HPR, HRG, HSD17B6, RDH16, SERPINC1, SLC10A1, SLC27A5, TAT, TTR, exosome-related genes AEBP1, ANTXR1, APLNR, ASPN, CRISPLD2, DCN, EFEMP1, FBLN2, FMOD, GGT5, LAMC3 , LUM, MFAP4, MGP, MXRA5, MYH11, SVEP1, and FTH gene FTH1) were used for molecular typing and survival risk assessment, and six reference genes (including GAPDH, GUSB, MRPL19, PSMC4, SF3A1, and TFRC) were used as internal standards. All 62 genes except the 6 reference gene...
Embodiment 3
[0150] Molecular classification and survival risk assessment of liver cancer by quantitative PCR
[0151] Experimental method: A 24-gene test combination was used, of which 21 liver cancer subtypes and survival risk-related gene groups (proliferation-related genes: BIRC5, CDC20, MKI67, MYBL2, TOP2A, TPX2 and UBE2C; immune-related genes: CD2, CD74 and CD8A; intercellular matrix-related genes: GLYAT, GYS2, HRG, SERPINC1, SLC10A1, TAT; exosome-related genes: DCN, EFEMP1, LUM and MFAP4; FTH gene: FTH1) for molecular typing and survival risk assessment, 3 A reference gene (ACTB, GAPDH and RPLPO) was used as an internal standard. All 21 genes in Table 2 except the reference gene were used to calculate the survival risk index.
[0152] Table 2: Molecular classification of HCC and / or genogroups for survival risk
[0153]
[0154]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com