Method for preparing circular ssDNA and kit of method
A kit and circular technology, applied in a method for preparing circular ssDNA and its kit, in the field of next-generation sequencing, can solve the problems of poor sequencing uniformity of high GC content samples, damaged library DNA strands, etc., and achieve improved sequencing Uniformity, the effect of improving the quality of sample sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
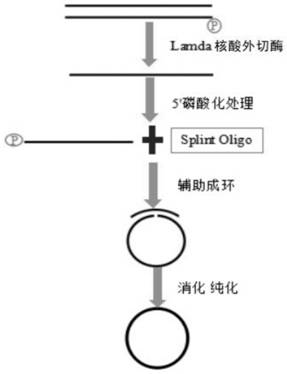
[0031] Example 1 Preparation of Halobacterium halophilus (GC%=64) circular ssDNA by the method of the present invention
[0032] 1. Synthesis of amplification primers:
[0033] According to the method for preparing circular ssDNA of the present invention, primers were synthesized, and primer Ad153_PCR 2-2 was subjected to 5' phos modification to obtain the primer pair as follows:
[0034] Ad153_PCR 2-1: GAACGACATGGCTACGA (SEQ No. 1)
[0035] Ad153_PCR 2-2: 5'-p-TGTGAGCCAAGGAGTTG (SEQ No.2)
[0036] 2. Construction of Halobacterium library (MGI platform):
[0037] A, sample selection: in this example, select halophilic bacilli with GC content of 64%;
[0038] B. DNA fragmentation: the sample gDNA is interrupted, and the fragmentation method selected in this example is the mechanical method;
[0039] C. Library construction: using Ultima DNA Library Prep Kit for formula kit, please refer to the instruction manual of the library construction kit for details; the amplifica...
Embodiment 2
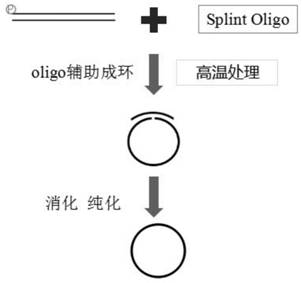
[0056] Example 2 High-throughput sequencing and result analysis
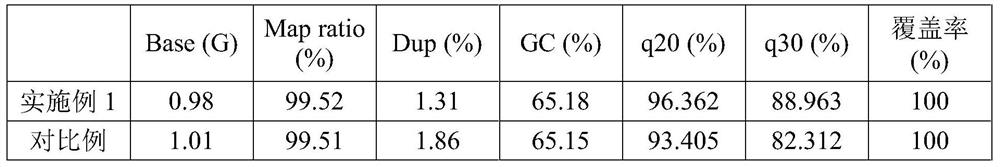
[0057] In this example, we used the halophilic library as a template, prepared single-stranded circular ssDNA by the above two circularization methods, and sent it to the MGI sequencing platform for sequencing in the same lane of the same sequencer. The sequencer is BGISEQ-2000, mode PE100, the amount of sequencing data is 1G; the quality of sequencing data is shown in Table 1 and Table 2.
[0058] Table 1 Different circularized ssDNA preparation methods - comparison of sequencing quality 1
[0059]
[0060] Table 2 Different circularized ssDNA preparation methods - comparison of sequencing quality 2
[0061]
[0062] The results show that the ssDNA library prepared by the method of the present invention has one of the important indicators for measuring the sequencing quality - the base quality value is higher than that of the ssDNA library prepared by the traditional method, and its Q20 and Q30 are incre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com