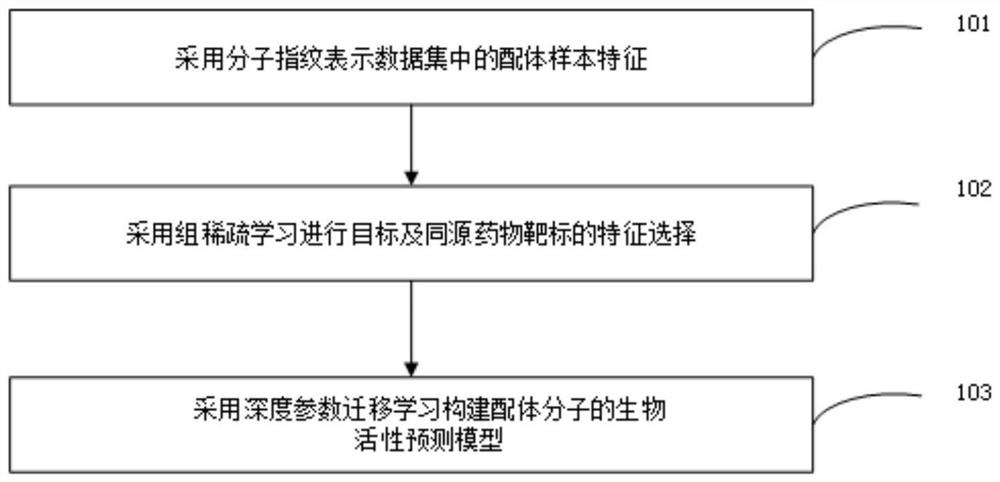
Small molecule drug virtual screening method and device based on deep parameter transfer learning
A transfer learning, deep parameter technology, applied in neural learning methods, molecular design, chemical machine learning, etc., can solve problems such as difficult to build virtual screening models
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
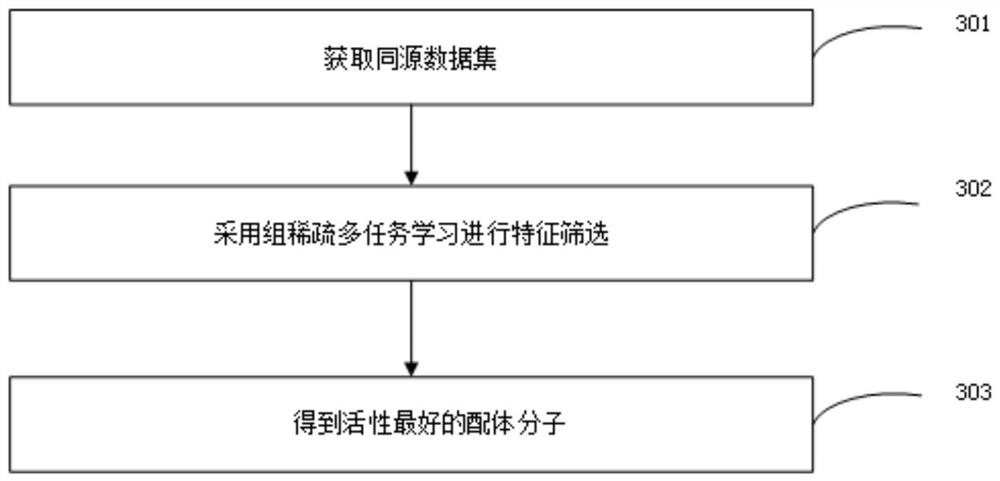
[0053] Using group-sparse multi-task learning to build a virtual screening model.
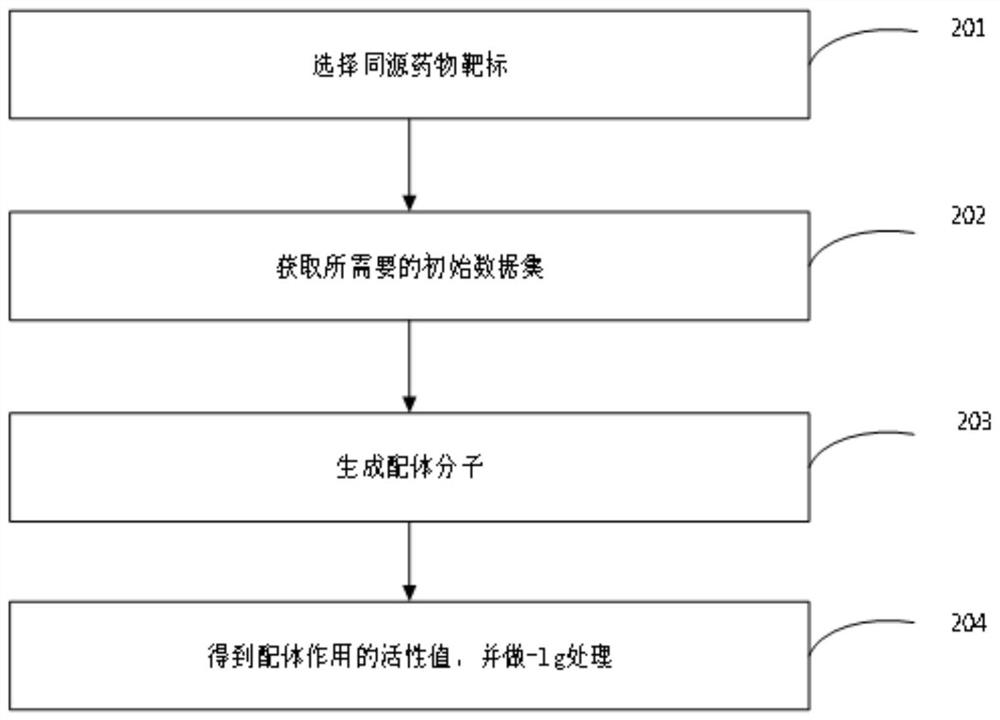
[0054] Select homologous drug targets, as shown in Table 1, Group B, including O95136, Q9H228, O95977, Q99500, and P21453, respectively numbered by B1, B2, B3, BS1, and BS2.
[0055] Table I
[0056]
[0057] Obtain the required initial dataset, including:
[0058] canonical smiles: the ligand of the ligand molecule, used to generate the molecular characteristics of the ligand;
[0059] standard value: the activity value of each ligand;
[0060] standard units: units.
[0061] Table II
[0062]
[0063] According to the initial data set, for each task, the ligand molecule is obtained, here is the smiles of each subject, and the corresponding characteristics are obtained by using the molecular fingerprint method;
[0064] Consider multiple tasks together, and take X=[x1…xn]T∈Rn×t as the input data feature matrix, denoted as X. The results are shown in Table 3.
[0065] Table three ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com