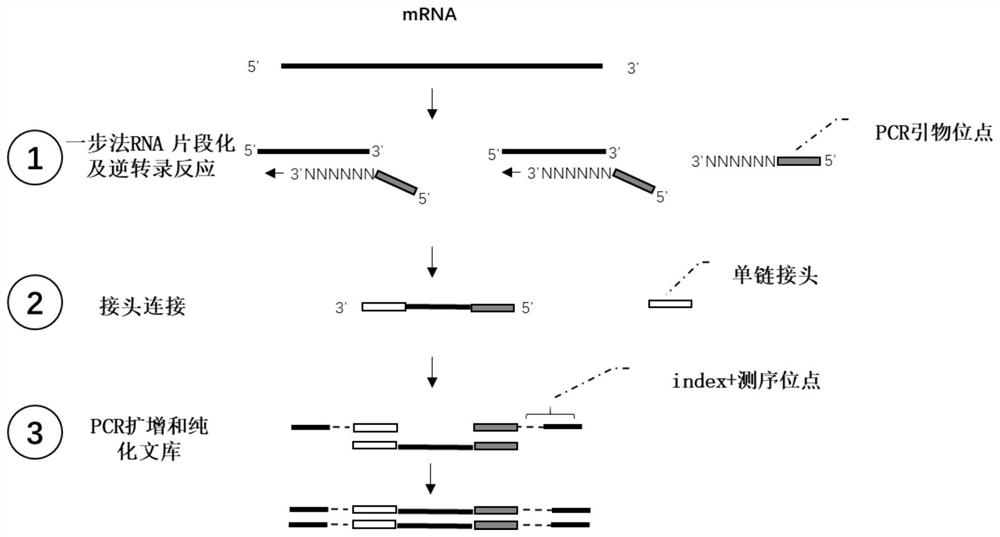
RNA library construction method
A construction method and library technology, applied in the field of RNA sequencing library construction, can solve problems such as cumbersome library construction process, and achieve the effect of simple operation and saving man-hours
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] 1. One-step method to complete RNA fragmentation and reverse transcription reaction (Novizan kit, product number: NR604)
[0049] (1) Use 19.5 μL fragmentation buffer (Fragment buffer) to elute the enriched RNA sample, heat at 94°C for 8 minutes to complete the RNA fragmentation reaction, and perform reverse transcription reaction immediately;
[0050] (2) Prepare the first-strand cDNA synthesis reaction system according to the following table:
[0051] Table 1:
[0052]
[0053] (3) Carry out the first-strand cDNA synthesis reaction in a PCR instrument:
[0054] Table 2:
[0055]
[0056] (4) Immediately purify with 1.8X DNA clean beads (Clean beads), then use 16μL ddH 2 O to elute the beads.
[0057] 2 single chain header connections
[0058] (1) configure the reaction solution by the following components:
[0059] table 3:
[0060]
[0061] Incubate at 60°C for 1 hour, and incubate at 80°C for 10 minutes to denature.
[0062] (2) Use magnetic beads t...
Embodiment 2
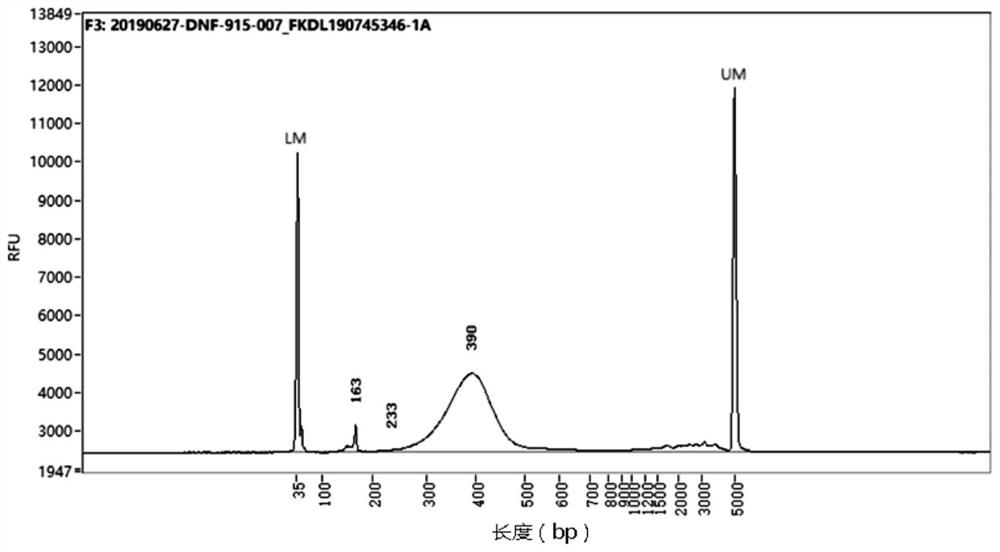
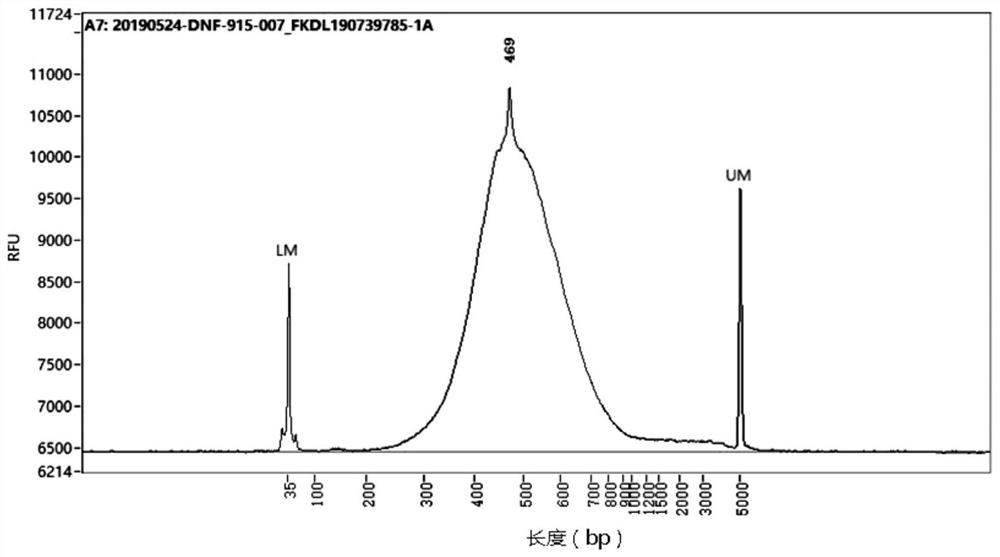
[0082] Compared with the common RNA-seq library construction method (that is, the method of building a library by synthesizing double-stranded cDNA mentioned in the background technology), using the method of the present invention in the test of tobacco samples, the quality inspection results of the libraries constructed by the two methods were respectively Such as Figure 3A with Figure 3B shown.
[0083] The analysis results of sequencing data are shown in Table 7. Q20 and Q30 are above 97% and 92% respectively, the mapping rate is above 94%, and other indicators meet the requirements. Figure 4 Shown are the difference graphs of gene expression detected after the libraries are constructed by the two methods, and it can be seen from the graphs that the results of the method of the present invention are consistent with those of the traditional method.
[0084] Table 7:
[0085]
Embodiment 3
[0087] Compared with the common RNA-seq library construction method, using the method of the present invention in the test of human samples, the quality inspection results of the libraries constructed by the two methods are as follows: Figure 5A with Figure 5B shown.
[0088] The analysis results of sequencing data are shown in Table 8. Q20 and Q30 are above 96% and 91% respectively, the mapping rate is above 95%, and other indicators meet the requirements. Image 6 Shown are the difference graphs of gene expression detected after the libraries are constructed by the two methods, and it can be seen from the graphs that the results of the method of the present invention are consistent with those of the traditional method.
[0089] Table 8:
[0090]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com