Device for noninvasive prenatal detection of chromosome abnormality
A technology for chromosomal abnormalities and prenatal detection, applied in the field of bioinformatics, can solve the problems of low efficiency, time-consuming, waste of sequencing costs, etc., and achieve the effect of reducing the number, improving the detection rate, and simplifying the function.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1 Fitting function establishment
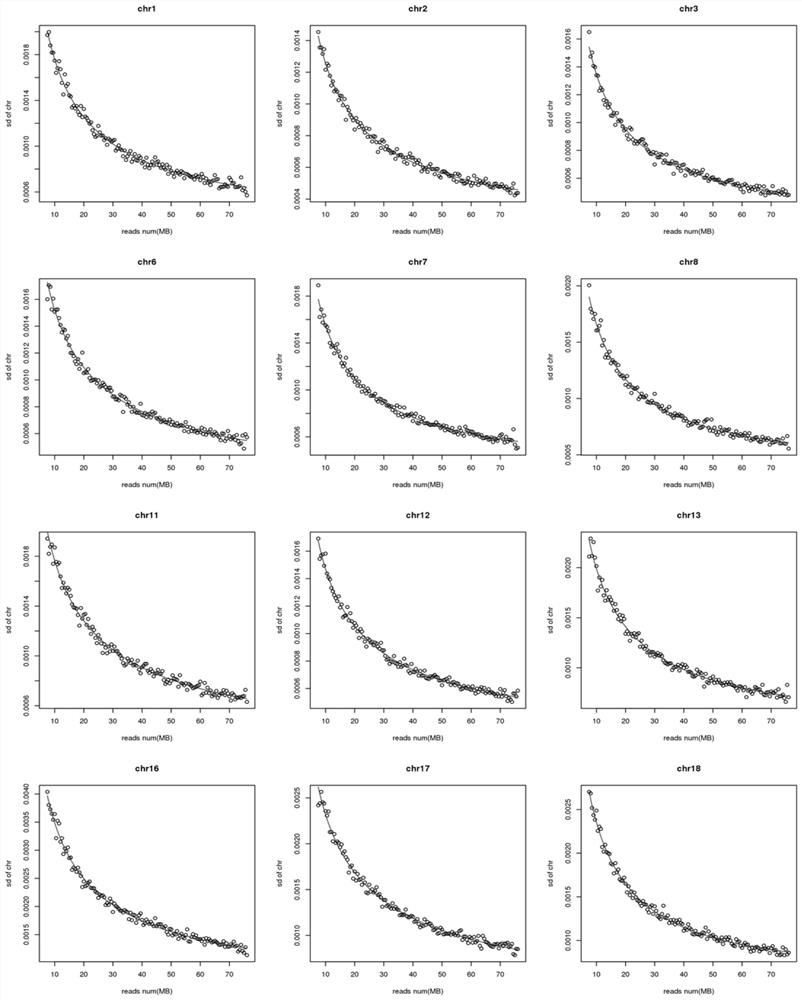
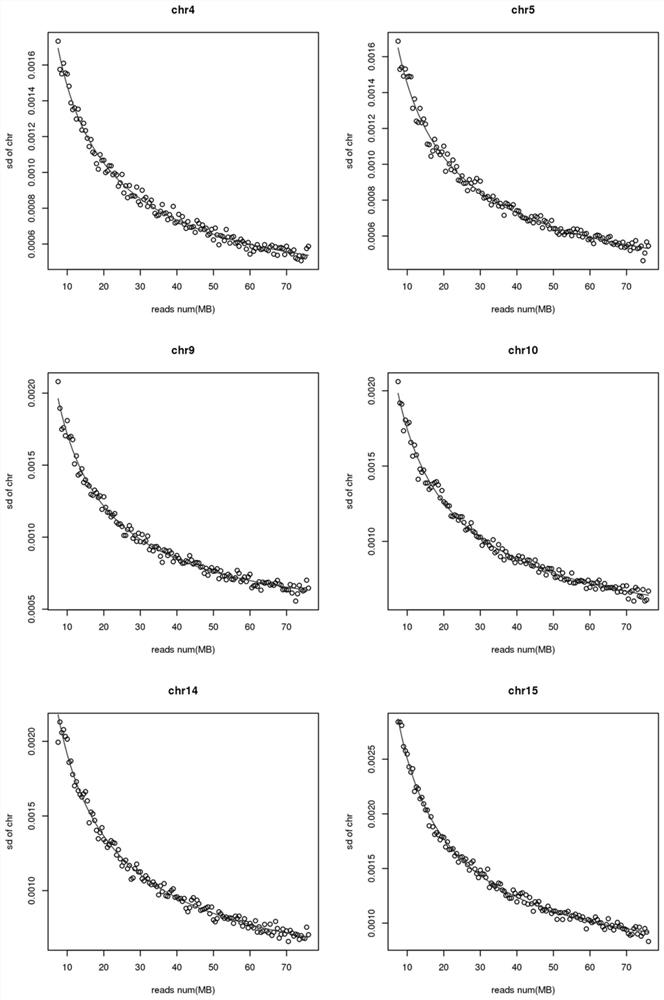
[0041] In this example, the standard deviation of the number of reads of 22 chromosomes at different sequencing depths is calculated. For the change diagram, see figure 2 , image 3 , Figure 4 .
[0042] Among them, each graph represents a chromosome, and the abscissa in each graph is the number of reads in the reference set sample, and the ordinate is the standard deviation of the chromosome under this number of reads. From Figure 2-Figure 4 It can be seen that the standard deviation of the number of chromosome reads at different sequencing depths changes significantly. As the number of reads increases (that is, the sequencing depth increases), the standard deviation will become smaller. This shows that for a sample, it should be used instead of The standard deviation parameter should be matched to the sequencing depth instead of using the same standard deviation for all samples.
[0043] In each figure, according t...
Embodiment 2
[0058] Embodiment 2 Chromosomal Abnormal Judgment
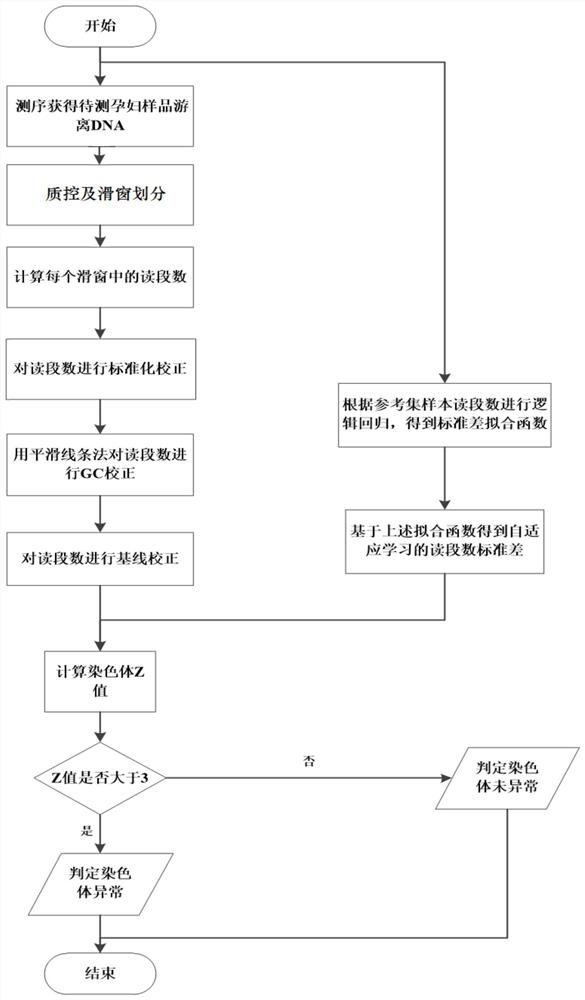
[0059] This embodiment provides a method for detecting chromosomal abnormalities using the device for non-invasive prenatal detection of chromosomal abnormalities of the present invention. For a schematic diagram of the testing process, see figure 1 . details as follows:
[0060] (1) Use the detection module to perform sequencing on the peripheral blood cell-free DNA data of the pregnant woman to be tested. The sequencing platform used is the MGI2000 platform. The fq file (including the sequencing sequence and the sequencing quality of the sequence) can be obtained through sequencing on the sequencing platform, and the read length is 50bp ;
[0061] (2) Use the data quality control module to perform quality control screening on the fq file: remove PCR repeats, remove low-quality reads containing consecutive N bases (or the average Phred score of 5 consecutive nucleotides is less than 20), if Remove the above-mentioned fra...
Embodiment 3
[0075] In this example, 135 true positive samples and 3 negative samples with chromosomal abnormalities were verified by using the method in Example 2 and the traditional method. The gestational weeks of the 138 samples were all greater than 10 weeks, and their peripheral blood data were taken for verification.
[0076] The detection process of the traditional method and the method of Example 2 of the present invention is only different in the process of calculating the Z value in the last step, and other processing processes including sequencing, calculating the number of reads in each window width, GC correction, baseline correction, etc. are all the same (as in Example 2 process).
[0077] Traditional method:
[0078] The inventive method:
[0079] As can be seen from the method of the Z value calculation of above-mentioned traditional method and the inventive method, the denominator of traditional method uses fixed standard deviation (in the present embodiment, the f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com