Reverse transcription obstruction probe for rapidly removing target RNA in RNA library construction and application thereof
A reverse transcription and probe technology, applied in the direction of DNA/RNA fragment, recombinant DNA technology, microbial determination/inspection, etc., can solve the problems that hinder the development of RNA next-generation sequencing technology, industrial automation development application, sample source and RNA abundance High requirements, difficult storage of reagents, etc., to achieve low cost, short time-consuming, and complete effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The features and advantages of the present invention can be further understood through the following detailed description in conjunction with the accompanying drawings. The examples provided are only illustrative of the method of the present invention and do not limit the rest of the present disclosure in any way. The sequences and modifications of the probes and primers used in this example are shown in Table 1. N is a random base, that is, any base in A, T, C, and G, and RandomHexamers is a universal primer for reverse transcription.
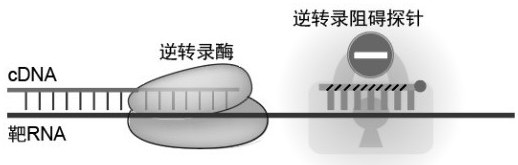
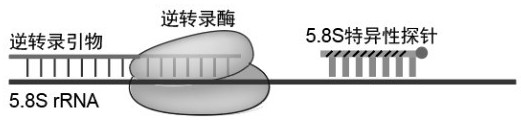
[0042] figure 1 and figure 2 The reaction mechanism of the present invention is demonstrated: the reverse transcription hindering probe (such as LNA, PNA, etc., and the 3' end of the probe is blocked by NH2C6) is modified with a base with strong binding ability, so that the reverse transcription hindering probe can preferentially lock the target RNA, preventing reverse transcriptase from blocking the target RNA site locked by the pro...
specific Embodiment approach
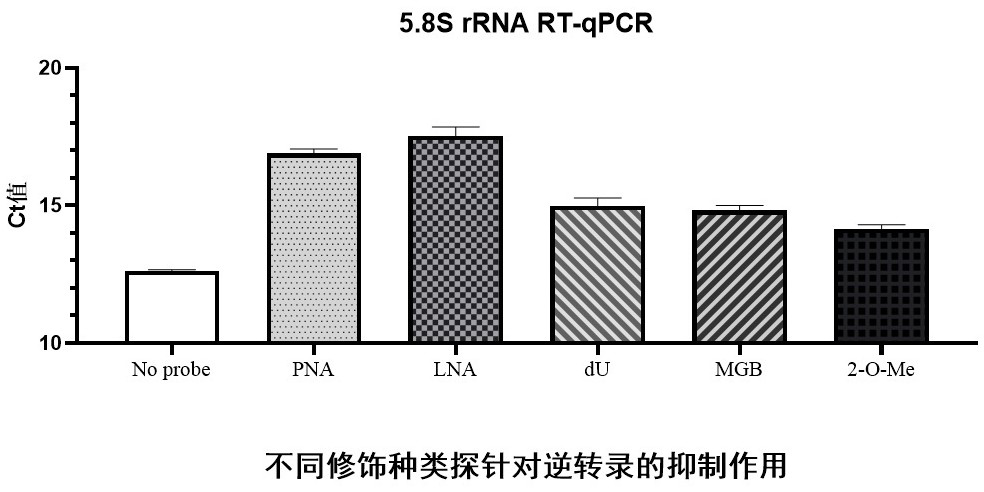
[0050] refer to figure 2 In this example, 5.8S-specific probes with different modifications (No. 1-5 in Table 1) were designed to inhibit the reverse transcription process of 5.8S rRNA. The Total RNA used in the examples was extracted from cultured Human293FT cells (GM-070006H, purchased from Jimon Biotechnology (Shanghai) Co., Ltd.) according to conventional methods, the same below. The specific implementation is as follows:
[0051] Table 2
[0052] components Dosage Total RNA 1 μg 1 μM 5.8S modified probe (any one of No. 1-5 in Table 1) 1 μL 1 μM 5.8S rRNA RT primer 1 μL 10 mM dNTPs mix 1 μL Supplement DEPC H2O to 13 μL
[0053] Mix and spin away. 85 °C for 5 min, then immediately place on ice for 5 min.
[0054] table 3
[0055] components Dosage 5X Reverse transcriptase buffer 4 μL Murine RNase Inhibitor (40 U / µL, Yeasen, 10603ES05) 2 μL Hifair® V Reverse Transcriptase (200 U / μL, Yeasen,...
Embodiment 2
[0061] Example 2: The inhibitory effect of the distribution position of LNA in the probe on the reverse transcription of target RNA
[0062] In this example, 5.8S-specific probes (numbers 6-8 in Table 1) of different LNA modification positions were designed to inhibit the reverse transcription process of 5.8SrRNA. With reference to example 1 for the specific embodiment, the quantitative results are shown in Figure 4 .
[0063] It can be seen from the quantitative results that when the position of the LNA in the probe is close to the left end of the probe (ie, the 5' end), the effect of hindering the reverse transcription of the target RNA is higher than that when the position of the LNA in the probe is close to the right end of the probe (ie, the 5' end of the probe). 3' end).
[0064] Table 5: Inhibition efficiency of target RNA reverse transcription by LNA distribution position in the probe
[0065] Distribution of LNA modification positions in probes Left ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com