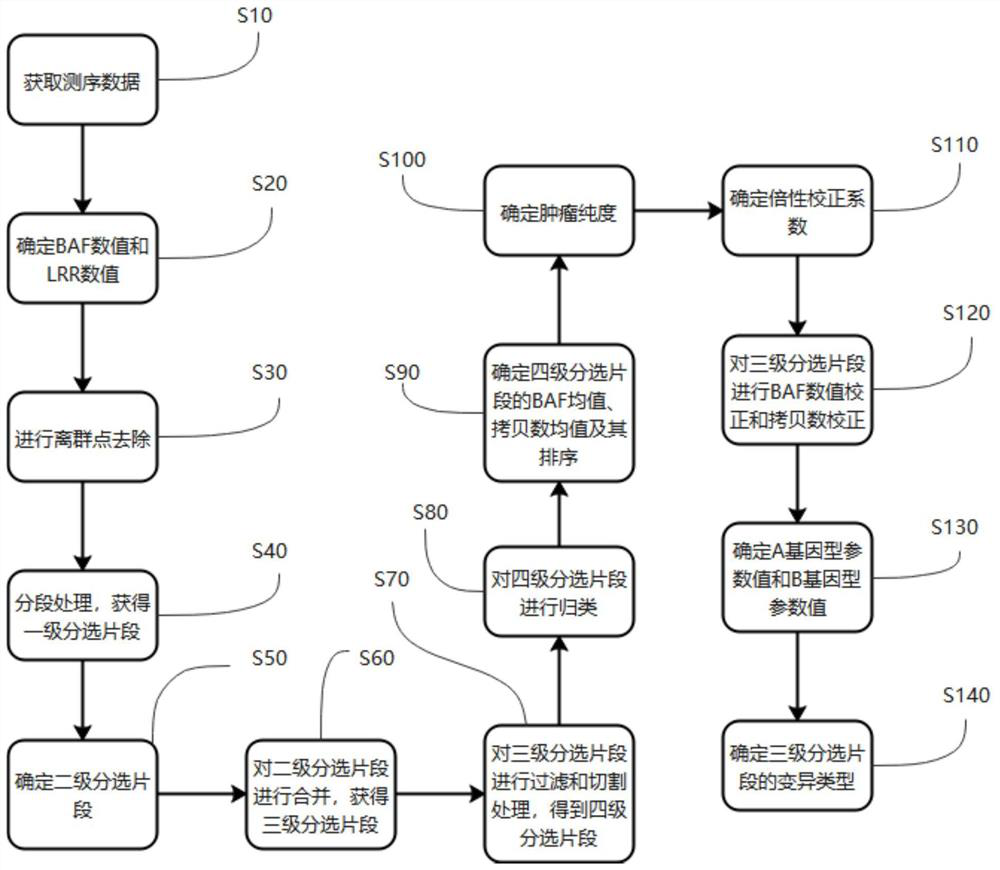
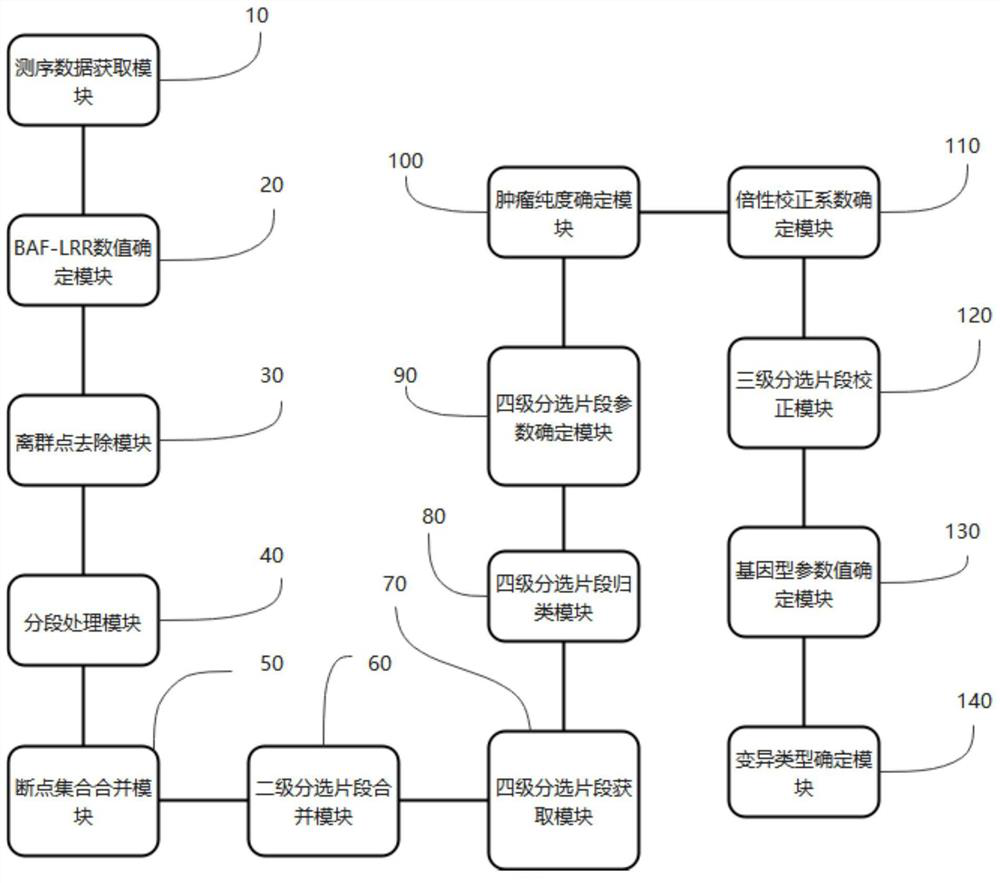
Kits, probes and applications for measuring genomic instability
A technology of instability and kits, applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems that the analysis of genome instability needs to be improved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0285] Example 1—Copy number detection and purity correction module in the method of the present invention (GSA method)
[0286] In order to reflect the optimization made by the inventor for the genotype detection of large fragments, the inventor selected the public software with better industry evaluation but no targeted optimization for comparison. PureCN is a relatively complete implementation of the ABSOLUTE algorithm, which is currently the mainstream method for purity correction based on copy number data. PureCN software updates are relatively active, and the inventor is currently using the stable version 1.8.1 for evaluation. In addition to the copy number calculation itself, the advantages and disadvantages of the method of the present invention and PureCN purity calculation are also an index in this comparison.
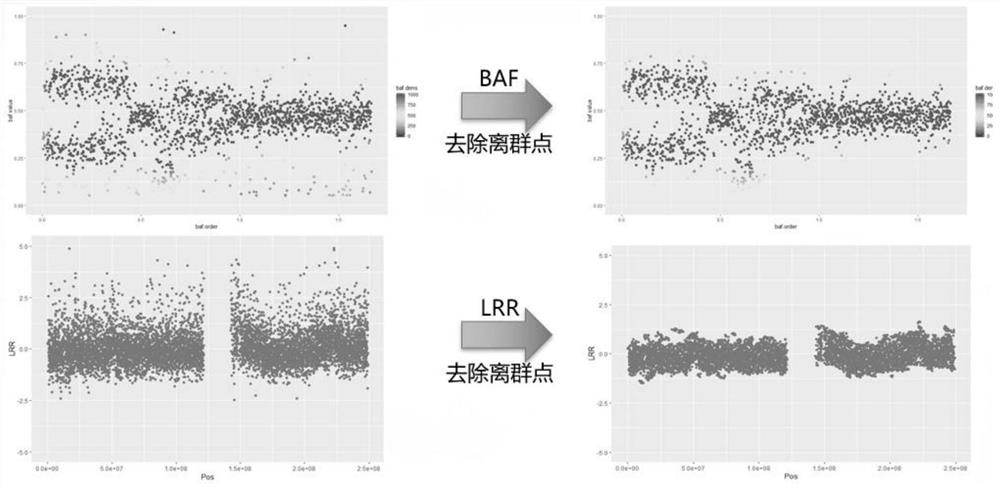
[0287]Here the inventors will show a scatter plot of BAF and LRR calculated for each capture interval. The paragraph division given by the software is repr...
specific Embodiment example 2
[0329] Specific implementation case 2—a kit for comprehensive evaluation of genomic instability and homologous homogeneous gene repair gene mutation detection
[0330] (1) Sample selection
[0331] Randomly select 3 samples of commercialized cell lines, and use the algorithm designed by the present invention and the kit for comprehensive evaluation of genomic instability and homologous homogeneous gene repair gene mutation detection to carry out the verification of the embodiment.
[0332] sample name HCC1395 HCC1954 MDA-MB-436
[0333] (2) Sample fragmentation
[0334] This kit is compatible with 100-400ng DNA samples, use nuclease-free ultrapure water (NF water) to make up its volume to 75μl. Transfer the liquid to the disruption tube and put it into the nucleic acid disruption instrument (Bioruptor pico). The interrupter is precooled to 4°C, and the interrupting parameters are:
[0335] Target Fragment Size (bp) Interrupt paramet...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com