Specific amplification primer pair and fluorescent quantitative PCR kit
A technology for amplification primers and fluorescent quantification, applied in the field of gene editing, can solve the problems of increasing false positives and being easily affected by external factors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Embodiment 1, design and synthesis of specific amplification primer pair
[0018] The GPCR gene was amplified, and analyzed and compared according to the LSDV GPCR gene sequence (accession number: MN508357) published in GenBank. A pair of specific primers were designed, the upstream primer qGPCR-F: 5'-AGTCGAATATAAAGTAATCAGTC-3', the downstream primer qGPCR-R: 5'-CCGCATATAATACAACTTATTATAG-3', and the length of the amplified fragment was 125bp. The above primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd.
Embodiment 2
[0019] Embodiment 2, the construction of positive standard
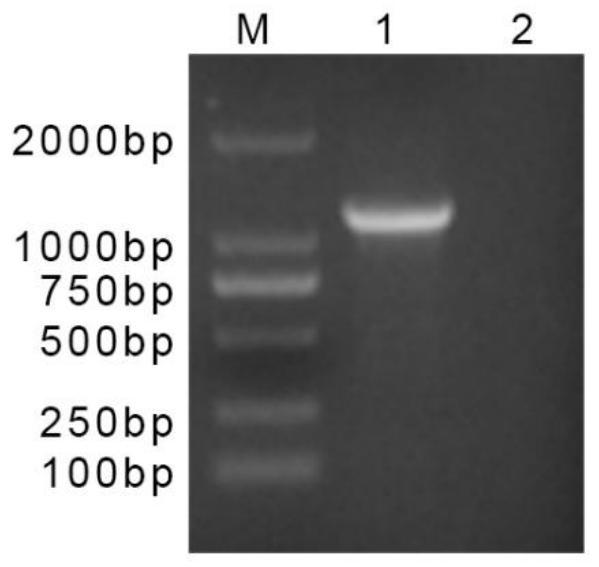
[0020] The DNA of LSDV was extracted according to the Mini BEST ViralRNA / DNA Extraction Kit, and its GPCR gene was amplified by ordinary PCR. The reaction system is 25 μL: 2×Premix Tap 12.5 μL, upstream and downstream primers 1.0 μL (10 μmol / L), template 1.0 μL, ddH 2 O 7.5 μL. The amplification program was 94°C for 5min; 94°C for 30s, 55°C for 30s, 72°C for 1min, 35 cycles; 72°C for 5min. The PCR amplification products were subjected to 1.0% agarose electrophoresis, such as figure 1 shown. Then follow the instructions of DNA Purification Kit to recover and purify the PCR product, and clone it into the pMD19-T vector to construct the recombinant plasmid pMD19T-LSDV, which is the positive standard. figure 1 Middle lane number 1 is the recombinant plasmid pMD19T-GPCR, and lane number 2 is the negative control.
[0021] The recombinant plasmid pMD19T-LSDV was sent to Shanghai Sangon Bioengineering Co., Ltd. for seq...
Embodiment 3
[0022] Embodiment 3, the establishment of fluorescent quantitative PCR standard curve
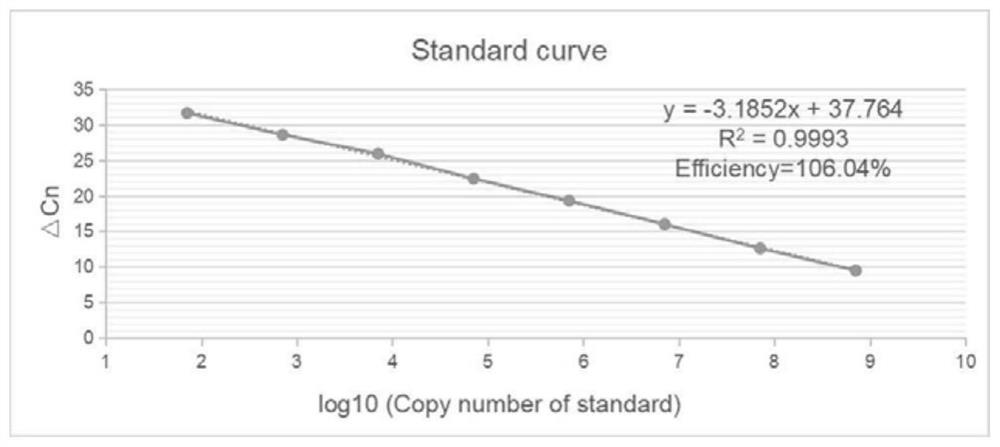
[0023] The recombinant plasmid pMD19T-LSDV was serially diluted to obtain 7.11×10 2 copies / μL~7.11×10 9 Copies / μL and other 8 dilution standards are used as the reaction template. Three samples were replicated for each dilution gradient and a negative control was established. Fluorescence quantitative PCR reaction system is 20 μL: TB Green (2×) 10 μL, upstream and downstream primers 0.8 μL (10 μmol / L), template 1 μL, ddH 2 O 7.4 μL. The reaction conditions were: 95°C for 30s, 1 cycle; 95°C for 10s; 60°C for 30s, a total of 40 cycles. Take the logarithm of the standard substance concentration as the X axis, and the Cq value as the Y axis, and draw a standard curve such as figure 2 As shown, the standard curve is y=-3.1852x+37.764, and the correlation coefficient R 2 =0.9993, the amplification efficiency Efficiency=106.04%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com