A library construction method for pacbio sequencing
A technology of sequencing library and construction method, applied in the field of library construction of PacBio sequencing, can solve the problems of loss of DNA samples, ineffective application, increased time consumption of sequencing libraries, etc., to improve the utilization rate and avoid the lower and lower circularization efficiency. Effect of total DNA requirement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1. Establishment of the PacBio sequencing library construction method based on Tn5 transposase
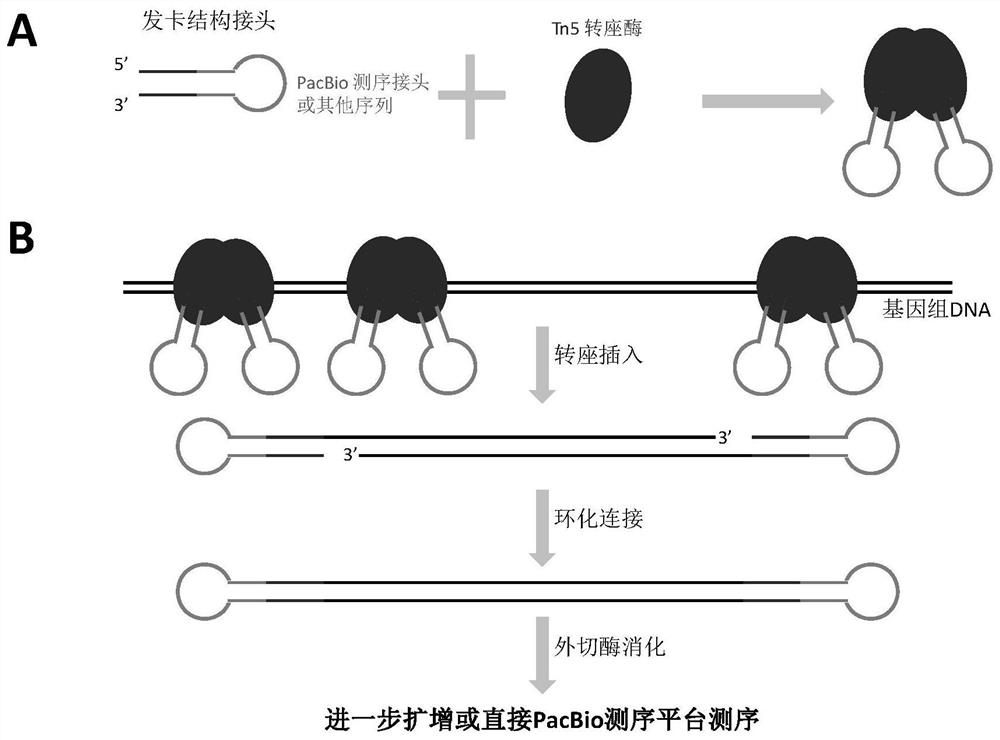
[0045] 1. Preparation of transposase complexes containing hairpin sequencing adapters ( figure 1 A)
[0046] 1. Design and synthesis of hairpin sequencing linker sequences
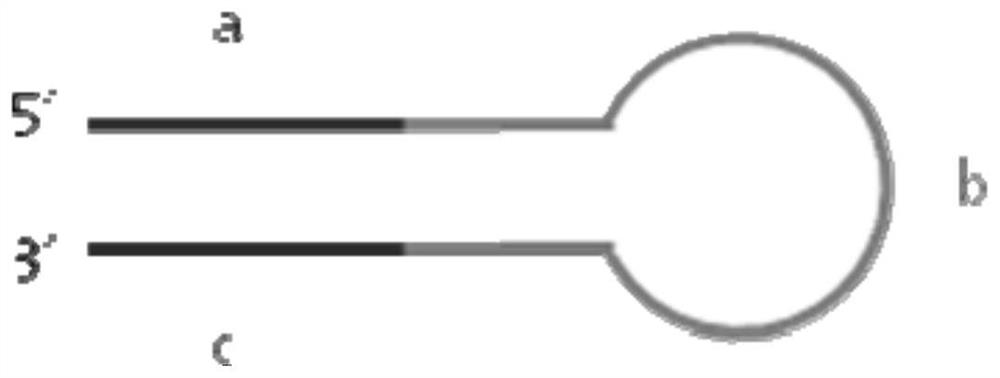
[0047] The sequence of the hairpin sequencing adapter is composed of fragment a, fragment b and fragment c sequentially from the 5' end ( figure 2 ), wherein fragment a and fragment c are completely reverse complementary.
[0048] Fragment a includes a transposase recognition sequence, and fragment a is phosphorylated from the first base at the 5' end;
[0049] In fragment a, a DNA barcode sequence for distinguishing different samples can also be introduced, and the DNA barcode is located at the 3' end of the transposase recognition sequence; the DNA barcode sequence is composed of more than 2 random bases, and the random bases are Any of ATCG;
[0050] Multiple samples correspond to multip...
Embodiment 2
[0070] Example 2, PacBio sequencing library construction based on constant (1 μg) DNA input of Tn5 transposase
[0071] 1. Preparation of the transposase complex PC0 containing the hairpin sequencing adapter ( figure 1 A)
[0072] 1. Design and synthesis of hairpin sequencing linker sequences
[0073] The artificially synthesized hairpin structure sequencing linker sequence PC0 is as follows:
[0074] PC0:
[0075] 5'-pCTGTCTCTTATACACATCT ATCTCTCTC TTTTCCTCCTCCTCCGTTGTTGTTGTT GAGAGAGAT AGATGTGTATAAGAGACAG-3' (SEQ ID NO: 1)
[0076] Among them, the 19 bases in bold at the 5' end ( figure 2 , Fragment a) can form a hairpin structure in a reverse complementary manner with the 19 bases (fragment c) that are thickened at the end of the 3' end; 19 bases at the end of the 5' end (fragment a), and 19 bases at the end of the 3' end The reverse complementary sequence of the base (fragment c) is the Tn5 transposase recognition sequence; the middle 45 base sequence (fragment b) ...
Embodiment 3
[0137] Example 3, PacBio sequencing library construction based on a small amount of (100ng) DNA input from Tn5 transposase
[0138] 1. Preparation of the transposase complex PC0 containing the hairpin sequencing adapter ( figure 1 A)
[0139] 1. Design and synthesis of hairpin sequencing linker sequences
[0140] The artificially synthesized hairpin structure sequencing linker sequence PC0 is as follows:
[0141] PC0:
[0142] 5'-pCTGTCTCTTATACACATCT ATCTCTCTC TTTTCCTCCTCCTCCGTTGTTGTTGTT GAGAGAGAT AGATGTGTATAAGAGACAG-3' (SEQ ID NO: 1)
[0143] Among them, the 19 bases in bold at the 5' end ( figure 2 , Fragment a) can form a hairpin structure in a reverse complementary manner with the 19 bases (fragment c) that are thickened at the end of the 3' end; 19 bases at the end of the 5' end (fragment a), and 19 bases at the end of the 3' end The reverse complementary sequence of the base (fragment c) is the Tn5 transposase recognition sequence; the middle 45 base sequence (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com