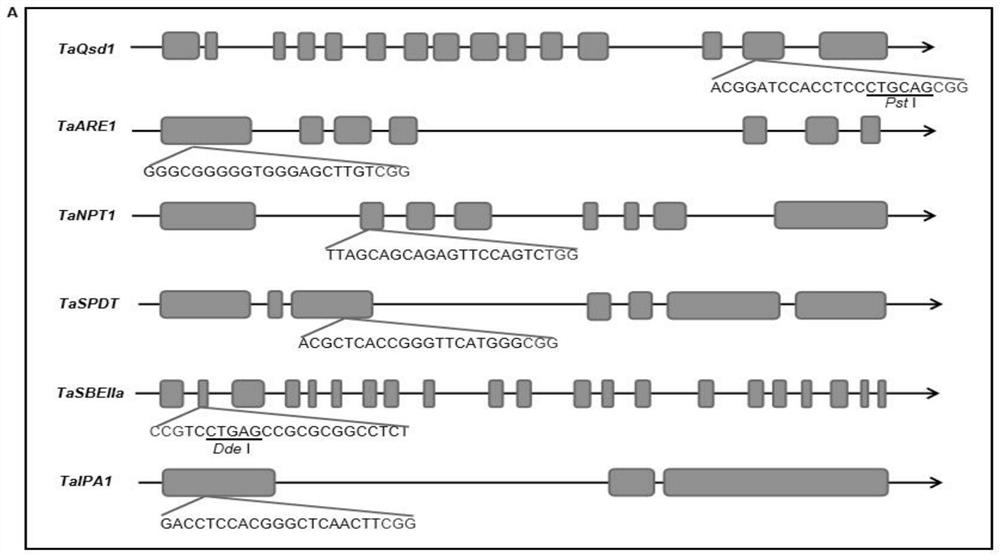
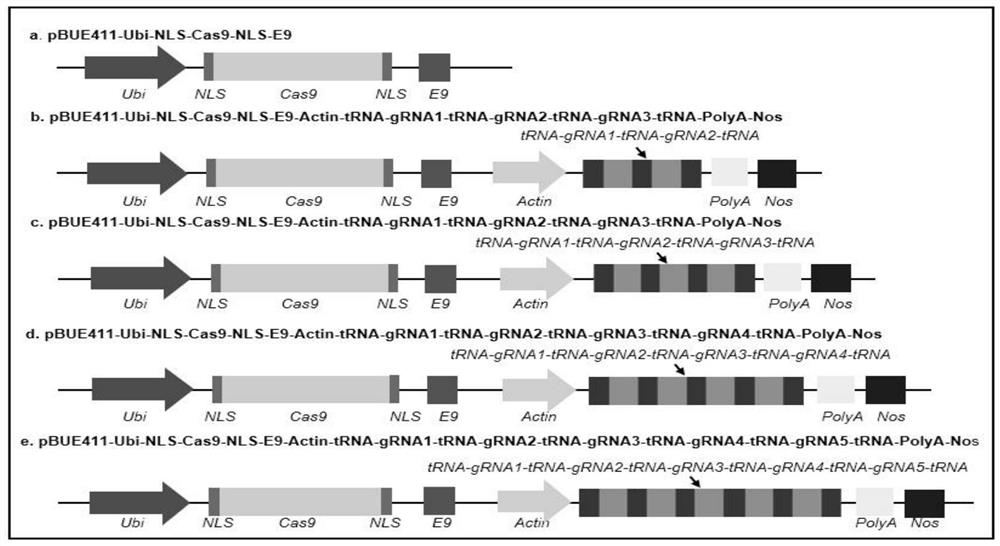
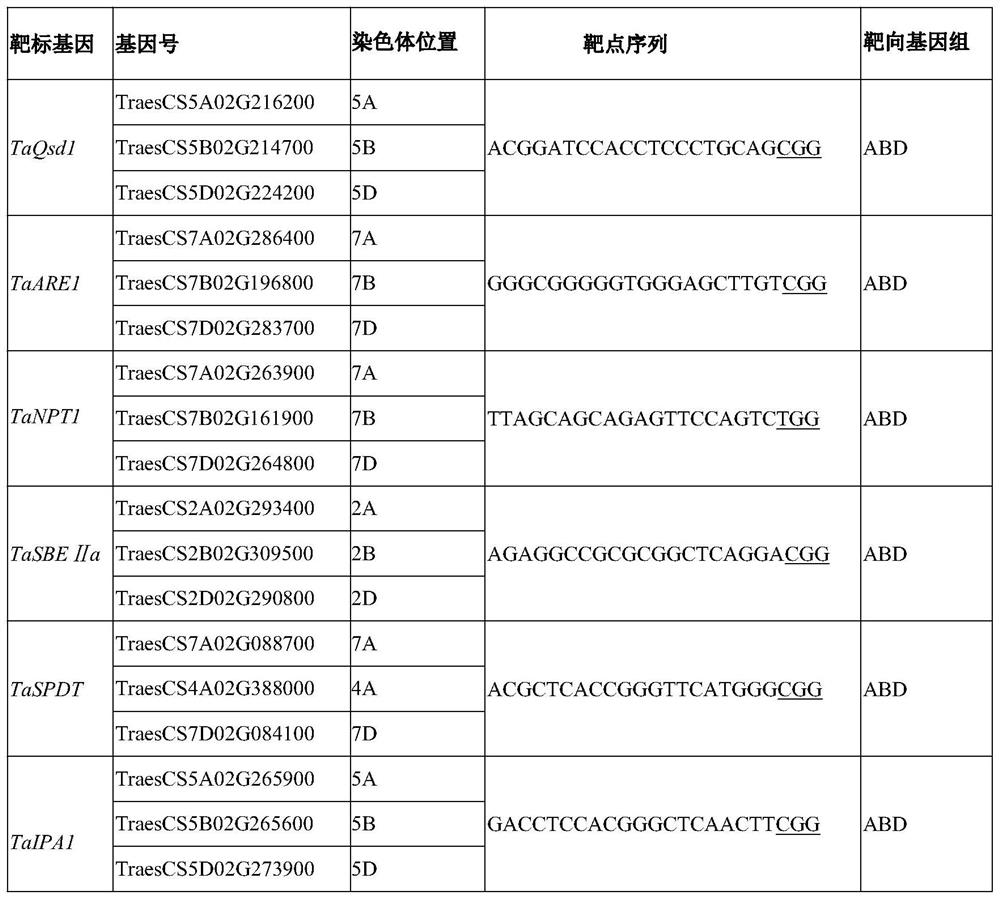
CRISPR/Cas9 system mediated wheat polygene knockout editing system
A multi-gene, gene technology, applied in genetic engineering, plant genetic improvement, angiosperm/flowering plants, etc., to achieve the effect of promoting research and speeding up complex traits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 Double-gene combination vector construction
[0053] 1.1 Construction of TaQsd1+TaNPT1 double gene combination knockout vector
[0054] First, the pBUE411-Ubi-NLS-Cas9-NLS-E9-Actin-PolyA-Nos vector was constructed on the pBUE411-Ubi-NLS-Cas9-NLS-E9 base vector. In the first round of PCR, two PCR systems were simultaneously amplified using the primer pair 414-M13-Actin-F / Nos-Hind-Act-R (Table 2) and Act-Hind-Nos-F / 414 -Ubi-Nos-R (Table 2), amplified with Actin sequence and Nos sequence as template, the second round of PCR using primer pair 414-M13-F / 414-Ubi-R (Table 2), with the first round of PCR The two PCR products obtained by PCR were used as templates to obtain the Actin-Nos expression cassette, and the expression cassette was digested with HindⅢ (NEB, Beijing, China) pBUE411-Ubi-NLS-Cas9-NLS-E9 basic vector, using the same Source recombinase (Quanshijin, Beijing, China) was connected to obtain the recombinant vector pBUE411-Ubi-NLS-Cas9-NLS-E9-Actin-Nos...
Embodiment 2 3
[0060] Example 2 Three-gene combination carrier construction
[0061] 2.1 Construction of TaQsd1+TaARE1+TaNPT1 triple gene combination knockout vector
[0062] Construction of pBUE411-Ubi-NLS-Cas9-NLS-E9-Actin-tRNA-gRNA1(TaQsd1)-tRNA-gRNA2(TaARE1)-tRNA-gRNA3(TaNPT1)-tRNA-PolyA-Nos three gene combination knockout vector. The first round of PCR uses tRNA as a template, and uses the primer pair 9-Actin-tRNA-F / 9-TaQsd1-tRNA-R to amplify, and the second round of PCR uses the tracrRNA-tRNA fragment as a template, respectively, using the primer pair 9-TaQsd1 -TrRNA-F / 9-TaAER1-tRNA-R and 9-TaARE1-TrRNA-F / PolyA-Hind-tRNA-R were amplified, and in the third round of PCR, the primer pair Actin-w-F0 / Poly- t-R0, using the three PCR products of the first round and the second round as templates to amplify to obtain the tRNA-gRNA1(TaQsd1)-tRNA-gRNA2(TaARE1)-tRNA fragment, which was then utilized by homologous recombinase ( Whole gold, Beijing, China) was cloned into the pBUE411-Ubi-Cas9-E9-A...
Embodiment 3 4
[0065] Example 3 Four-gene combination vector construction
[0066] 3.1 Construction of TaQsd1+TaARE1+TaNPT1+TaIPA1 Four-gene Combination Knockout Vector
[0067] The first round of PCR uses the tracrRNA-tRNA fragment as a template, and uses primers to amplify tRNA-TaIPA-F0 / PolyA-Hind-tRNA-R, and the second round of PCR uses the first round of PCR product as a template, using primers tRNA-TaIPA -F / PolyA-Hind-tRNA-R was amplified to obtain the gRNA4(TaIPA1)-tRNA fragment, which was digested with pBUE411-Ubi-NLS-Cas9-NLS-E9 with HindⅢ (NEB, Beijing, China) -Actin-tRNA-gRNA1(TaQsd1)-tRNA-gRNA2(TaARE1)-tRNA-gRNA3(TaNPT1)-tRNA-PolyA-Nos vector, using homologous recombinase (Quanshijin, Beijing, China) to obtain TaQsd1+ TaARE1+TaNPT1+TaIPA1 four-gene combination knockout vector pBUE411-Ubi-NLS-Cas9-NLS-E9-Actin-tRNA-gRNA1(TaQsd1)-tRNA-gRNA2(TaARE1)-tRNA-gRNA3(TaNPT1)-tRNA-gRNA4( TaIPA1)-tRNA-PolyA-Nos. The sequence of the TaQsd1+TaARE1+TaNPT1+TaIPA1 four-gene combination knockout...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com