Sic1 gene knock-in saccharomyces cerevisiae genetically engineered bacterium as well as construction method and application thereof
A technology of genetically engineered bacteria and Saccharomyces cerevisiae, applied in the field of genetically engineered bacteria of Saccharomyces cerevisiae and its construction, can solve the problems of prolonged fermentation period, high resistance, hindering mass transfer and conduction, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Construction of recombinant bacteria S288c-pSic1.
[0048] 1. Construction of the Sic1 gene knock-in module
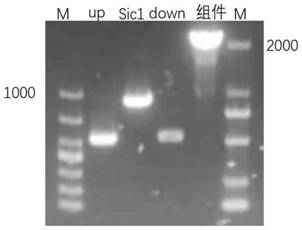
[0049] (1) Using the genomic DNA of the original Saccharomyces cerevisiae S288c as a template, the upstream, Sic1 gene and downstream homology arm amplification fragments were amplified by common PCR (upstream homology arm amplification primer sequences up-F, up -R are respectively shown in SEQ ID NO.2 and SEQ ID NO.3; Sic1 gene amplification primer sequences Sic1-F, Sic1-R are respectively shown in SEQ ID NO.4 and SEQ ID NO.5; downstream homology The sequences of the arm amplification primers down-F and down-R are respectively shown in SEQ ID NO.6 and SEQ ID NO.7); the PCR reaction system is shown in Table 1, and the PCR reaction conditions are as follows: 1) Pre-denaturation at 95°C for 3 minutes; 2 ) Denaturation at 95°C for 15 s, annealing at 60°C for 15 s, extension at 72°C for 1 min, three steps for 35 cycles, and extension at 72°C for 5 min. ...
Embodiment 2
[0094] (1) Take 10 μL of glycerol bacteria S288c (original bacteria) and S288c-pSic1 (knock-in bacteria constructed in Example 1) and add them to 5 mL of sterilized LYPD liquid medium for overnight culture and activation;
[0095] (2) According to the inoculation ratio of 1% by volume, transfer the bacterial liquid obtained in step (1) to 100 mL of YPD liquid medium, and continue to cultivate at 30 ° C and 200 r / min until the OD600 of the bacterial liquid is about 2.0;
[0096] (3) Take 2 mL of the bacterial solution obtained in step (2) and measure the absorbance value at OD600, and dilute the bacterial solution with sterilized YPD liquid medium so that the OD600 of the diluted bacterial solution is 1;
[0097] (4) Add 190 μL of YPD medium and 10 μL of the diluted bacterial solution in step (3) to a 96-well plate, and incubate at 30°C for 24 hours;
[0098] (5) Pour out the 96-well plate bacterial solution, buffer with 0.01M PBS buffer 3 times, and pat dry;
[0099] (6) Add ...
Embodiment 3
[0102] (1) Take 10 μL of glycerol bacteria S288c (original bacteria) and S288c-pSic1 (knock-in bacteria constructed in Example 1) and add them to 5 mL of sterilized LYPD liquid medium for overnight culture and activation;
[0103] (2) According to the inoculation ratio of 1% by volume, transfer the bacterial liquid obtained in step (1) to 100 mL of YPD liquid medium, and continue to cultivate at 30 ° C and 200 r / min until the OD600 of the bacterial liquid is between 0.8 and 1.2 ;
[0104] (3) Transfer the seed solution in step (2) to 100 mL of fermentation medium at an inoculation volume ratio of 10%, and divide it into free fermentation and immobilized fermentation.
[0105] When carrying out immobilized fermentation: add cotton fiber material to the fermentation medium as the immobilized material, add 4g cotton fiber medium to each shake flask; ferment at 30°C, 200r / min, after the glucose is exhausted, the reaction ends The instrument measures the amount of residual sugar i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com