An optimized acceptor splice site module for biological and biotechnological applications
A splice site, receptor technology, applied in the field of new receptor splicing regions, can solve the problem of low in vivo efficiency of rAAV dual vector system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0703] Example 1: Identification of optimized ASS modules
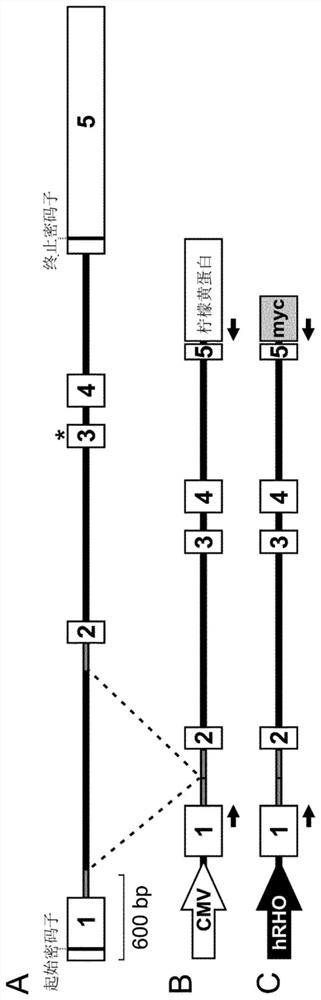
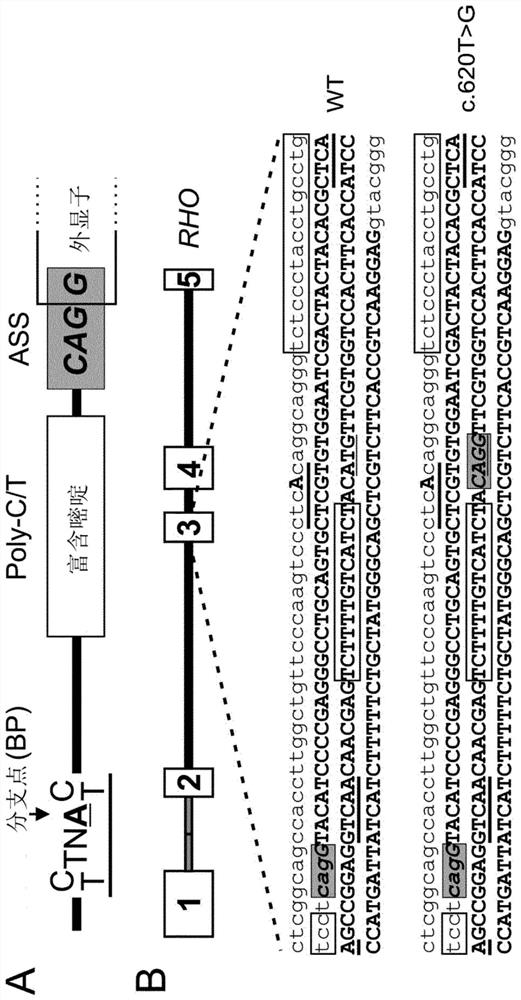
[0704] The effect of disease-associated rhodopsin mutations on mRNA splicing was analyzed using the human rhodopsin (RHO) minigene in HEK293 cells and transduced murine photoreceptor cells. Among them, we found a mutation (c.620T>G) in exon 3 of the RHO gene, which produces a new ASS ( figure 1 ). The sequence of ASS and predicted ASS elements as figure 2 A and B are shown.
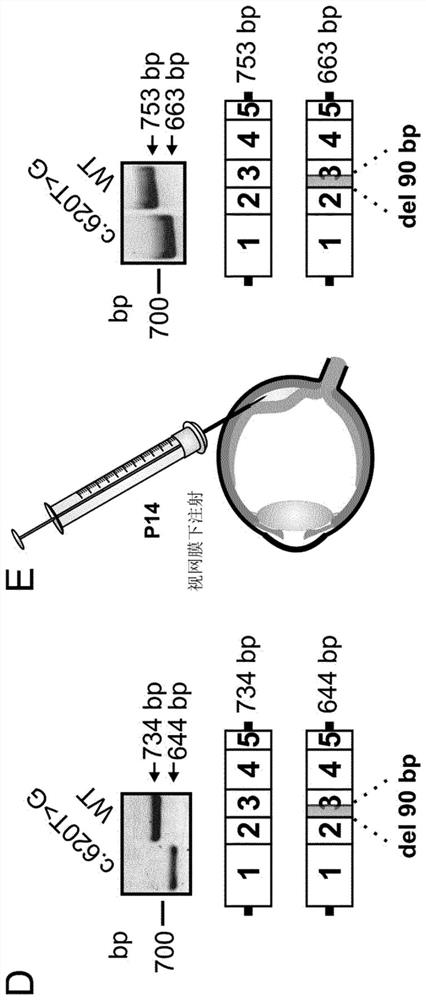
[0705]In silico predictions revealed that the receptor splice site formed by the c.620T>G mutation (hereinafter referred to as ASS_620) has a similar splicing fraction to the native rhodopsin receptor splice site in exon 3 (see image 3 A). This suggests that the splicing machinery can alternately use the two splice sites. However, experimental data suggest that ASS_620 is exclusively for HEK293 cells ( figure 1 D) and mouse photoreceptors ( figure 1 E). This suggests that ASS_620 is a strong acceptor splice site.
[0706] For potential...
Embodiment 2
[0708] Example 2: Cerulein reporter assay for in vitro applications
[0709] To test this hypothesis, a splicing reporter assay was generated by splitting the coding sequence of azurein into two artificial exons interrupted by an artificial intron ( Figure 4 D). The intron contains vgASS_620 and a strong donor splice site, separated from each other by an artificial 145bp. Confocal imaging of transfected HEK293 cells reveals strong and robust azurein fluorescence when both splice sites are presented in cis ( Figure 4 F(c1)). RT-PCR experiments and sequencing of the corresponding bands confirmed that, in this configuration, the two azure exons are efficiently spliced together (data not shown). Such as Figure 4 As shown in G, a specific immune signal of the expected size (27 kDa) was detected by Western blot using an antibody specific for the N-terminal portion of azurin. Taken together, these findings suggest that reporter assays are functional and highly efficient in ...
Embodiment 3
[0725] Example 3: Effect of acceptor splice site versus binding domain (BD) on remodeling efficiency.
[0726] BD length and sequence represent key determinants of remodeling efficiency. BDs most likely affect the tight binding and potential folding of mRNAs, but they are not expected to directly contribute to the efficiency or precision of the subsequent splicing process. As mentioned above, DSS is well characterized and predictions of its strength match well with its experimental performance. Therefore, there is no apparent need to optimize this splice site within the framework of split fluorophore remodeling assays. In contrast, the strength of ASS cannot be reliably predicted due to its complexity. The results showed that vgASS_620 is a particularly strong receptor splice site. Considering that splice site strength can affect remodeling efficiency for split fluorophore assays, vgASS_620 should result in higher values compared to other acceptor splice sites.
[0727] To...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com