Cancer driver gene prediction method and system based on local and global network centrality analysis
A technology of driving genes and prediction methods, applied in the fields of genomics, instrumentation, proteomics, etc., to achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0049] The preferred embodiments of the present invention will be described in detail below in conjunction with the accompanying drawings, so that the advantages and features of the present invention can be more easily understood by those skilled in the art, so as to define the protection scope of the present invention more clearly.
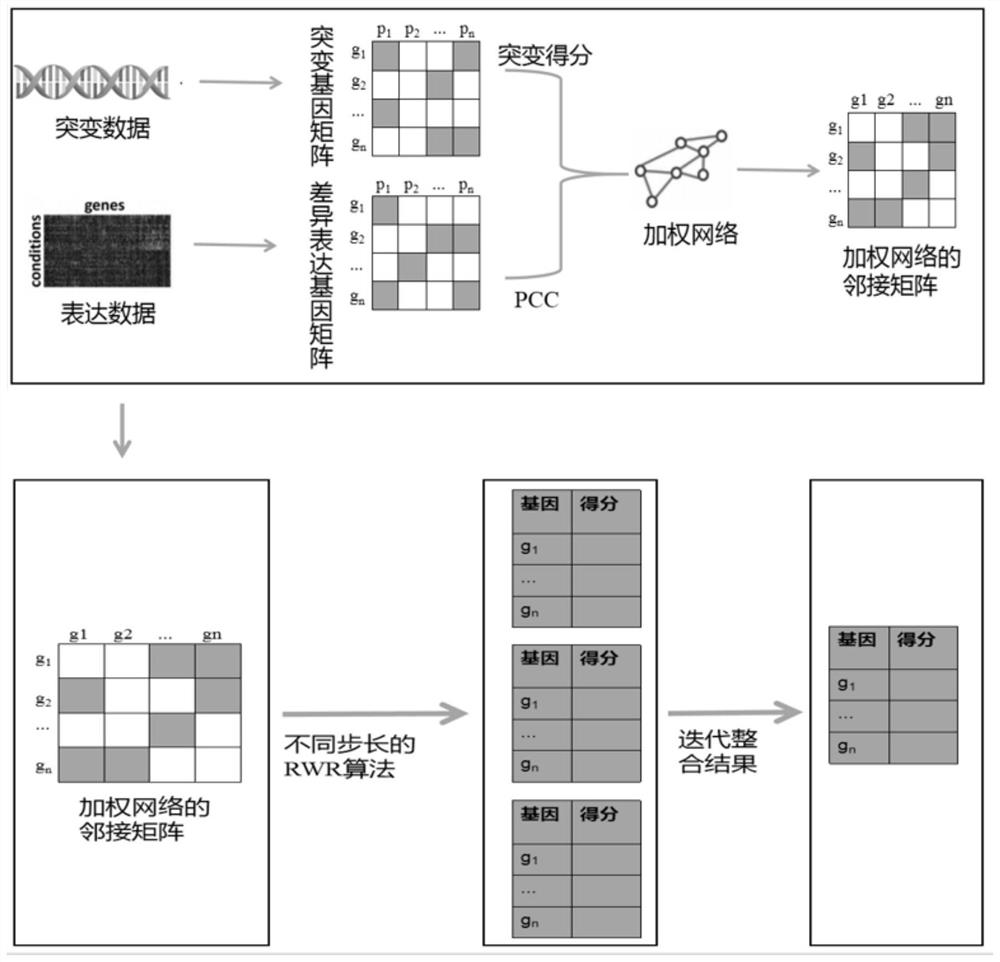
[0050] see figure 1 , the embodiment of the present invention includes:
[0051] A cancer driver gene prediction method based on local and global network centrality analysis, comprising the following steps:
[0052] S1: Preprocess the standardized somatic mutation data and gene expression data and express them in the form of gene-matrix;
[0053] Preferably, normalized somatic mutation data and gene expression data are downloaded from The Cancer Genome Atlas (TCGA) database. Further, the somatic mutation data include single nucleotide variations (SNVs) and chromosome aberrations (CNVs). Get validated cancer genes as positive samples (i.e. driver...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com