A detection method and quality control system for homologous recombination defects based on ngs platform
A detection value and sequencing technology, applied in the field of bioinformatics analysis, can solve problems such as accuracy limitations, lower sample HRD scores, and affect the accuracy of clinical sample HRD test results, etc., to achieve corrected accuracy, good detection limit and sequencing depth Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
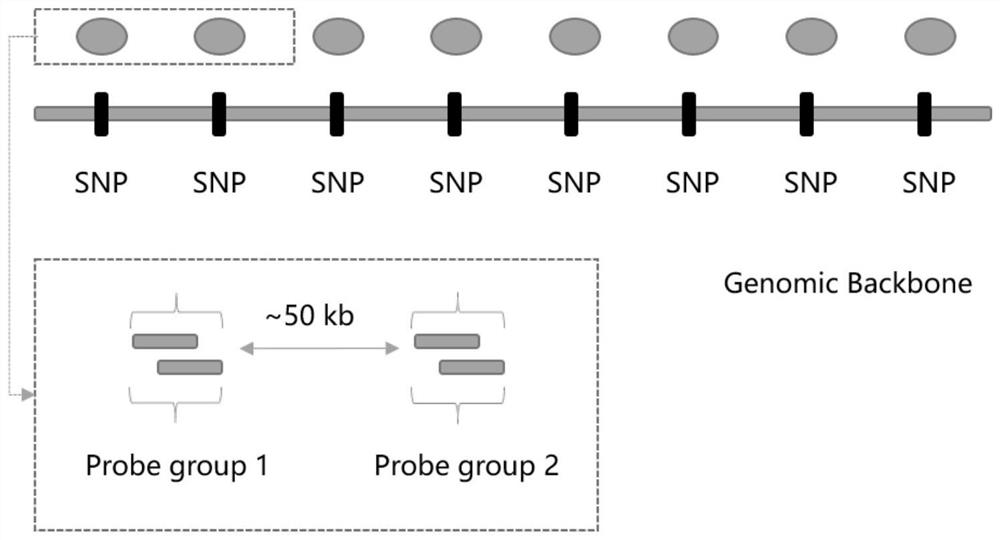
[0137] The Panel design method in this embodiment is as follows, exemplary, as shown in FIG. 1 .
[0141] Through the above design, 54,000 Backbone and 80,000 SNP sites were finally obtained.
Embodiment 2
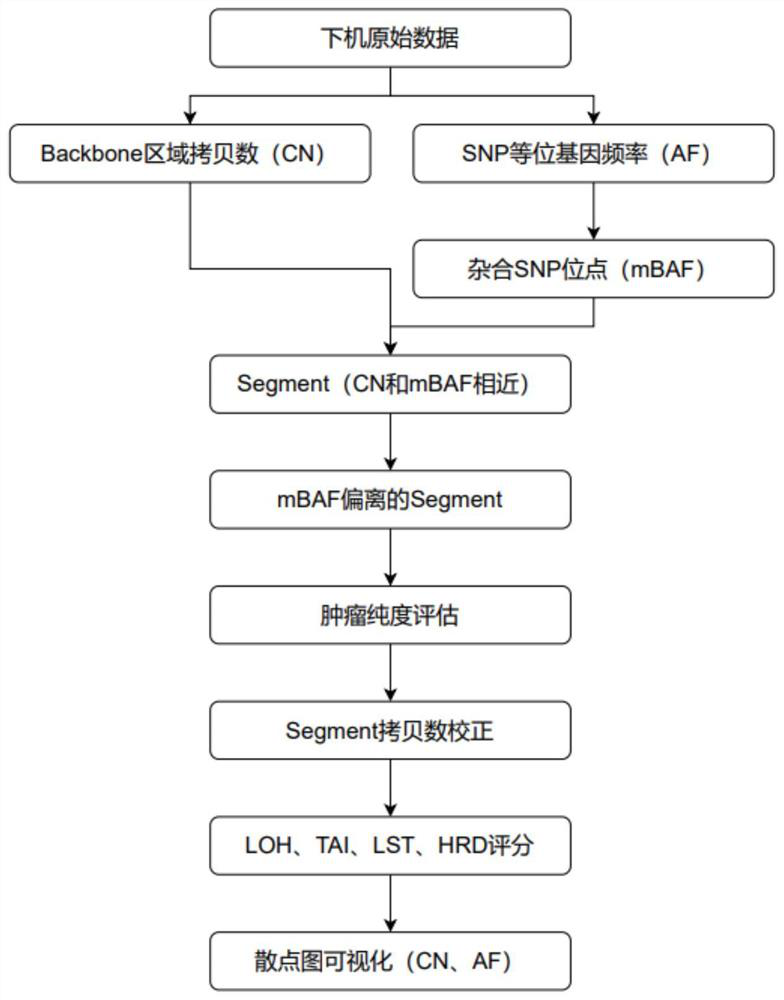
[0149] mBAF=|AF-0.5|+0.5
[0156]
[0160]
[0163] 9) Visualization, plotting AF scatter plots of Backbone region copy numbers and SNPs.
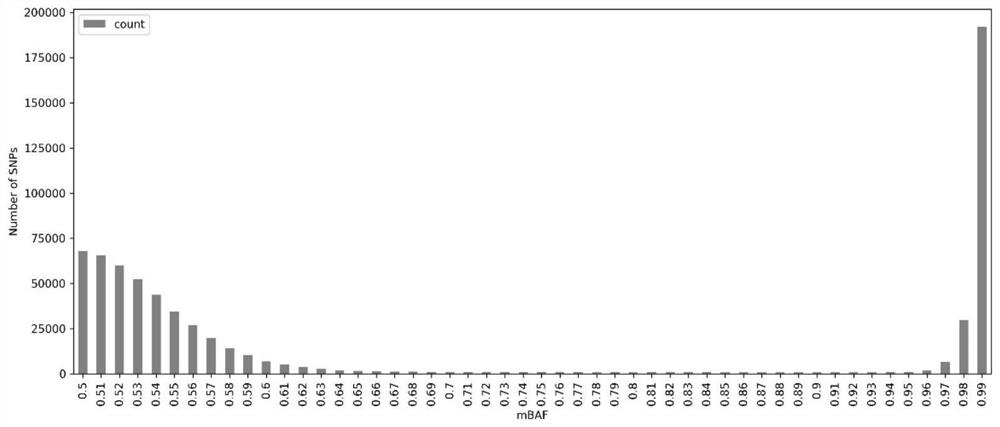
[0166] All SNP loci were heterozygous (AF=0.5, mBAF) due to the absence of copy number variation in negative samples
[0169]
[0172] Moreover, as shown in Figures 6-9, when the contamination ratio is less than or equal to 5%, adjusting the mBAF threshold (0.90) can make the same
Embodiment 3
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com