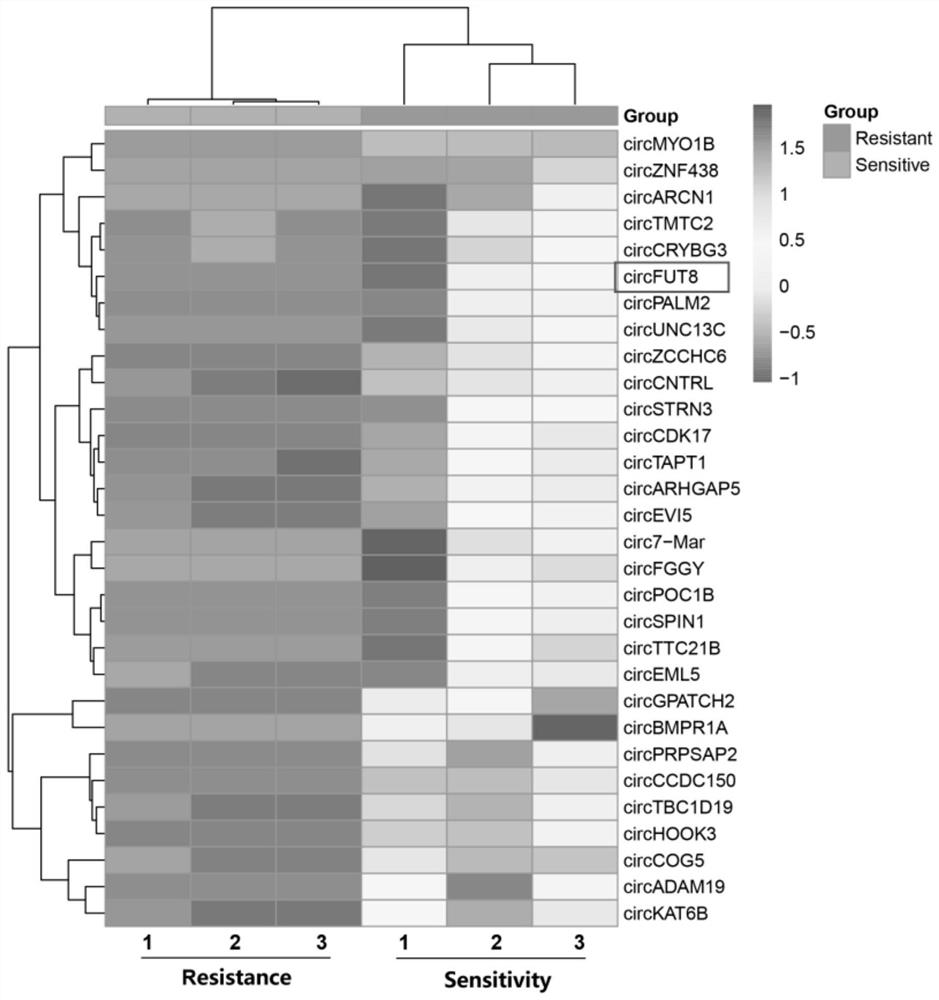
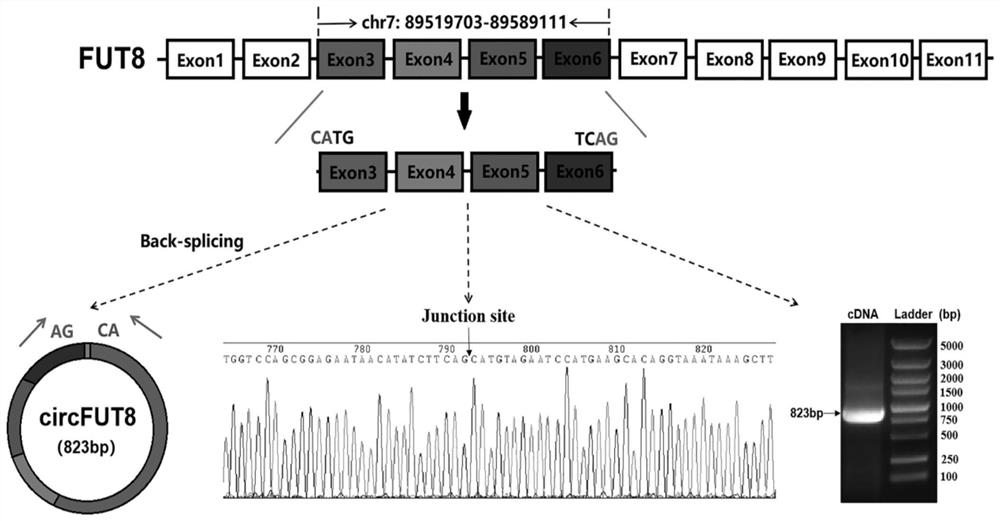
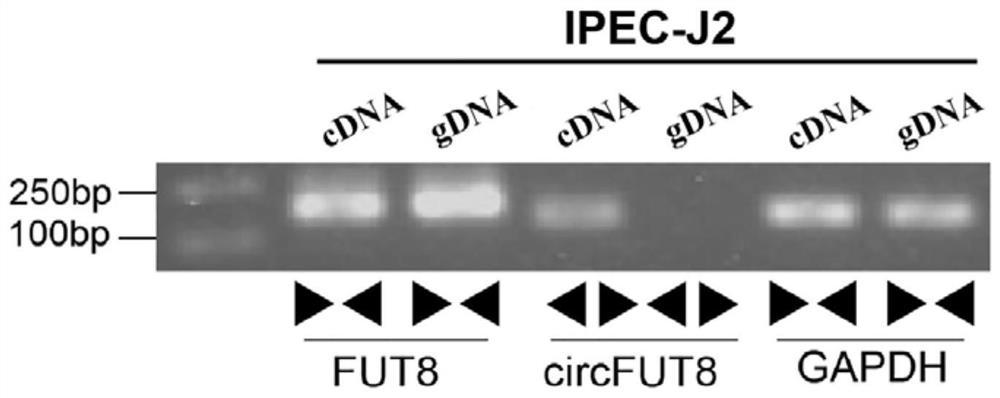
Pig circRNA sequence, application thereof and cyclization identification method
A sequence and looping technology, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problem of less research on porcine circRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0037] 1. Materials and methods
[0038] 1.1 RNA and genomic DNA (gDNA) extraction
[0039] In this study, the Trizol extraction kit (Invitrogen, USA) was used to extract total RNA from porcine small intestinal epithelial cells (IPEC-J2). The cells were lysed by adding the lysate according to the instructions, and then the binding solution was added to mix and pass through the column. The column was washed twice, and the RNA was finally eluted and tested for RNA purity, concentration, and integrity. Genomic DNA (gDNA) extraction was performed according to the instructions of the kit (Sangon Biotech, China).
[0040] 1.2 RNase R treatment, cDNA synthesis and PCR amplification
[0041] The same aliquot of RNA (2 μg) samples was incubated with 3 U / μg RNase R (Epicenter Technologies, USA) for 30 minutes at 37 °C, and then processed by RNeasy MinElute purification kit (Qiagen, Germany). Operate according to the instructions of the reverse transcription kit, and use RNA as a templa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com