Method and kit for detecting nucleic acid sequence of target to be detected by using melting curve of fluorescent probe
A nucleic acid sequence and melting curve technology, applied in the field of molecular biology, can solve the problems of low discrimination, non-specific Tm peak, low difference, etc., and achieve the effect of ensuring detection sensitivity, obvious difference in Tm value, and reducing detection cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] The basic principle of embodiment one inventive method
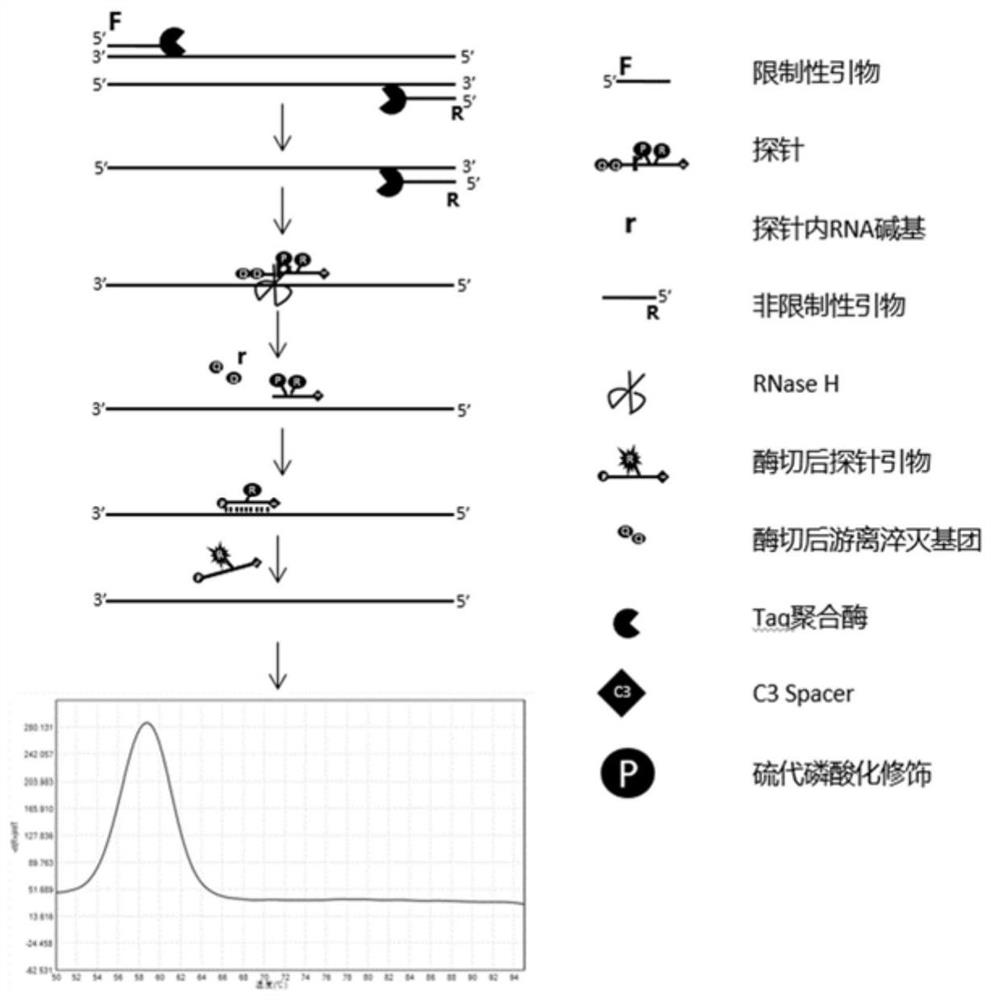
[0046] Such as figure 1 as shown in figure 1 As shown, the present invention utilizes the melting point Tm value of the fragment containing the fluorescent group on the right side of the probe RNA base in the fluorescent quantitative PCR amplification to bind to the product to realize single-tube detection of multiple target nucleic acid sequences and / or sequences in the sequence. SNP detection:
[0047] Step 1: Design specific upstream and downstream primers and probes for each target nucleic acid sequence to be tested, and realize the control of the melting point Tm value of the amplification product of each target nucleic acid sequence to be tested and the probe-binding fragment, so that the same fluorescence The amplification products of different target nucleic acid sequences to be detected in the labeling channel have differences in the melting point Tm values of the fragments formed by the combination o...
Embodiment 2
[0052] Example 2: Comparative experiment of labeling single / double quencher probes
[0053] Taking the detection of respiratory virus pathogen HCoV-229E as an example, use the method of the present invention but use respectively labeled single / double quencher probe to carry out qualitative detection to the RNA that pseudovirus extracts, and specific method comprises the following steps:
[0054] 1. Extraction of pseudovirus RNA
[0055] Use the viral RNA extraction kit of Tiangen Biochemical Technology (Beijing) Co., Ltd. to extract the viral RNA in the pseudovirus sample according to the instructions.
[0056] 2. Design of primers and probes
[0057] Design upstream and downstream primers and probes with one RNA base according to the conserved region of the nucleic acid sequence to be tested, where P1 is a labeled single-quencher probe, and P2 is a labeled double-quencher probe. The sequence information of the primer probes is shown in the table below .
[0058]
[0059...
Embodiment 3
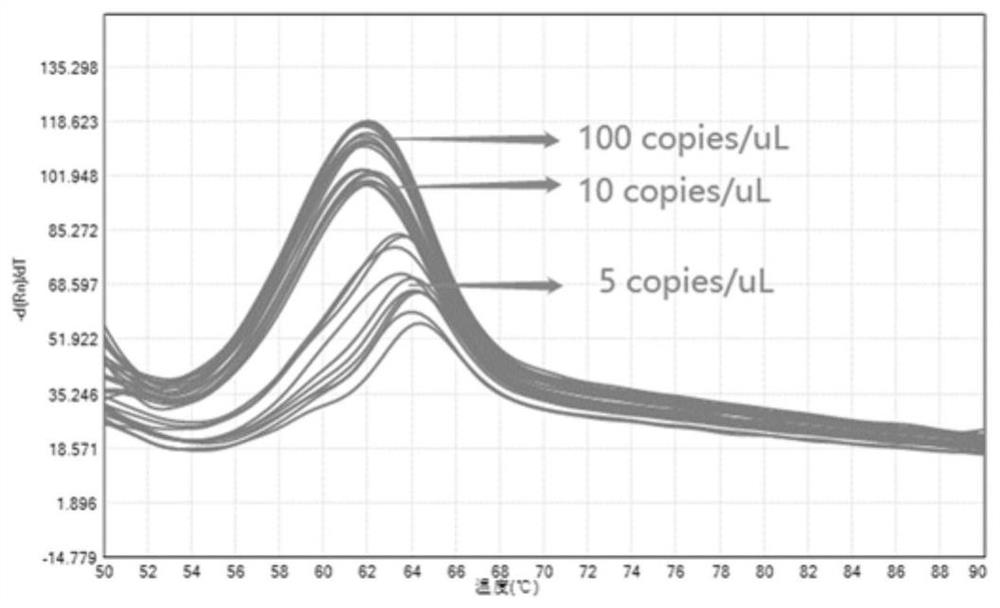
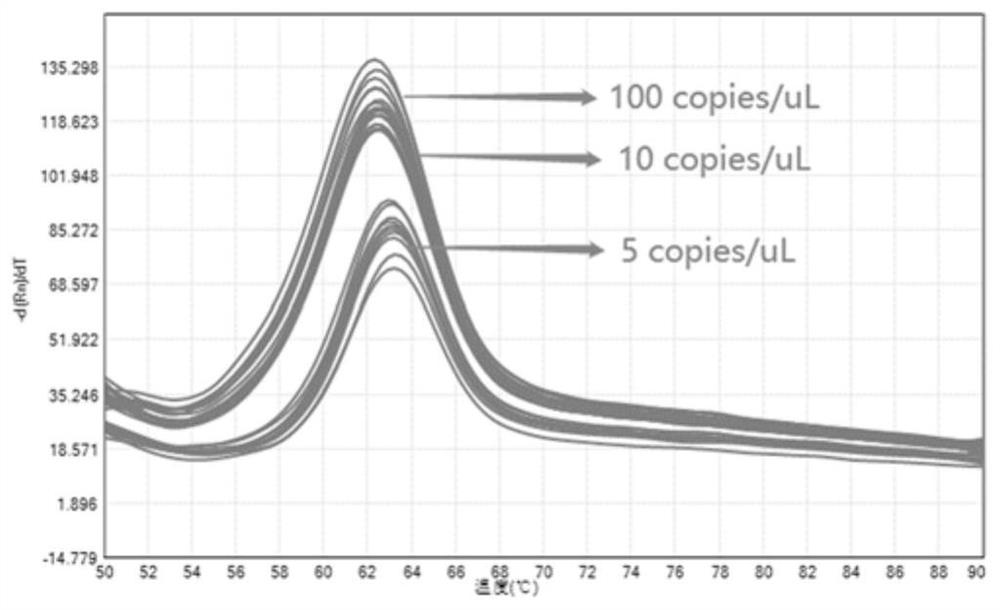
[0067] Example 3: Establishment of six-fold detection kit for respiratory virus pathogens
[0068] Take respiratory virus pathogen: novel coronavirus, HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1 and influenza B (IFB) detection are example, use the method of the present invention to carry out qualitative to the RNA that each pathogen pseudovirus extracts Detection, and qualitative detection of RNA in clinically positive samples of the 2019 novel coronavirus, HCoV-229E, HCoV-OC43 and influenza B (IFB) four pathogens. The specific method includes the following steps:
[0069] 1. Pseudovirus and RNA extraction from clinical samples
[0070] Use the viral RNA extraction kit of Tiangen Biochemical Technology (Beijing) Co., Ltd. to extract pseudoviruses and viral RNAs in clinical samples according to the instructions.
[0071] 2. Design of primers and probes
[0072] Design upstream and downstream primers and probes with one RNA base according to the conserved regions of each nucle...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com