Compositions and methods for treating alpha-1 antitrypsin deficiency
A technology for antitrypsin and deficiency, applied in biochemical equipment and methods, other methods of inserting foreign genetic materials, gene therapy, etc., can solve gene therapy that increases liver A1AT, cannot fully solve A1AT gene defects, cannot solve problems such as liver toxicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[1203] Example 1: Evolved adenosine base editors with increased editing efficiency
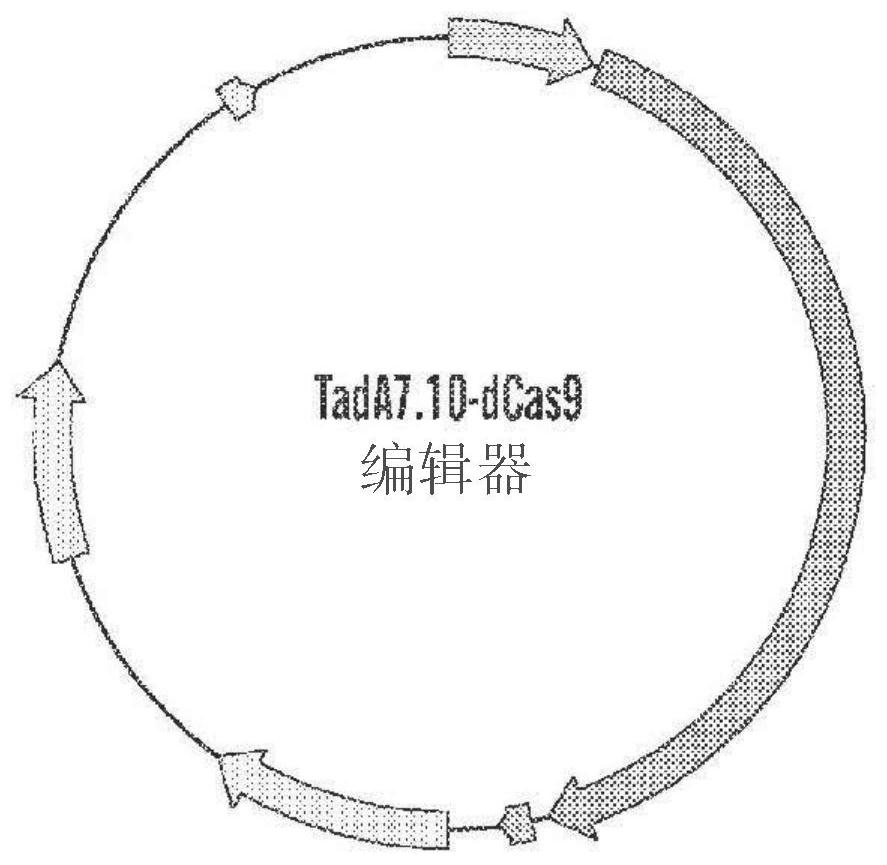
[1204] Base editing systems comprising Tad7.10-dCas9 fusion proteins are capable of editing target polynucleotides with approximately 10-20% efficiency, but their use may be limited for applications requiring higher efficiencies. To identify adenine base editors with improved efficiency and specificity, a construct containing the adenosine deaminase TadA 7.10 was mutagenized by error-prone PCR and subsequently cloned into a DNA-binding protein encoding dCas9 (nucleic acid-programmable DNA-binding protein). In the expression vector adjacent to the nucleic acid sequence ( Figure 1A ). The mutagenized TadA-dCas9 base editor is referred to in these Examples as ABE8 (adenosine deaminase variant). Expression vectors containing adenosine deaminase variants were co-transformed into competent bacterial cells with selection plasmids encoding chloramphenicol resistance (CamR) and spectinomycin resista...
Embodiment 2
[1213] Example 2: Correction of the alpha-1 antitrypsin mutation using ABE8
[1214] Base editing activity of selected ABE8 constructs was tested in HEK293 cells expressing A1AT containing the E342K mutation (HEK293T-E342K). In one scheme, 250ng gRNA plasmid and 750ng ABE8 plasmid [0407] were used to transiently transfect HEK293T-E342K cells with Mirus TransIT293, a highly efficient and low-toxic DNA transfection reagent optimized for HEK293 cells, at a ratio of 3 μl:1 μg. HEK293T-E342K was transfected by Neon electroporation using 2.5ug Var-3 ABE mRNA and 1000ng of 20nt gRNA 191. The gRNA backbone of the sgRNA provided as spCas9 base editor is as follows:
[1215] 5'-GUUUUAGAGC UAGAAAUAGC AAGUUAAAAU AAGGCUAGUC CGUUAUCAAC UUGAAAAAGUGGCACCGAGU CGGUGCUUUU-3'
[1216] Useful gRNAs for the method include the following
[1217] 5’-ACCAUCGACAAGAAAGGGACUGA GUUUUAGAGC UAGAAAUAGC AAGUUAAAAUAAGGCUAGUC CGUUAUCAAC UUGAAAAAGU GGCACCGAGU CGGUGCUUUU-3’
[1218] 5'-CCAUCGACAAGAAAGGGACUGA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com