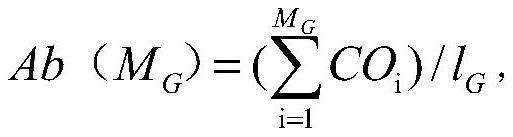Graves disease marker gene and application thereof
A technology for Graves' disease and marker genes, applied in the field of Graves' disease marker genes and their applications, can solve problems such as difficult differential diagnosis, Parkinson's disease confusion, complexity and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1 Identification of biomarkers
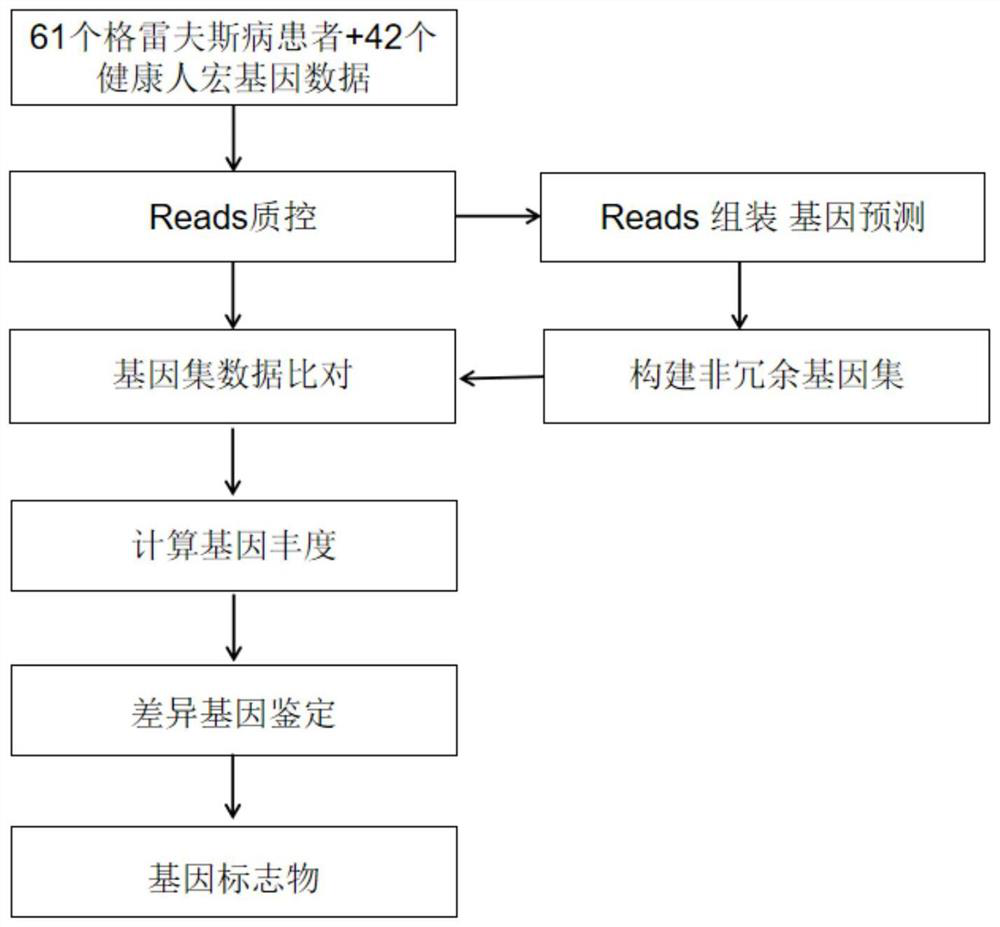
[0074] In this example, the inventors studied the fecal samples of 61 Graves' disease patients and 42 healthy controls to obtain the characteristics of the intestinal flora gene community and functional components. In general, the inventor downloaded about 353.3Gb of high-quality sequencing data (healthy people) and 513.9Gb of high-quality sequencing data obtained from experimental sequencing to construct a reference gene set for Graves' disease. Metagenome analysis showed that 44 genes were closely related to Graves' disease, 41 of which were enriched in the gut genes of healthy people, and 3 genes were enriched in the gut genes of Graves' disease patients.
[0075] 1. Acquisition of sequencing data
[0076] All sample sequencing data were downloaded from literature (PMID: 34079079) from Hainan Provincial People's Hospital, Haikou, China, aged 24-69 years, each subject in the healthy group collected stool samples before the fir...
Embodiment 2
[0102] Verification of embodiment 2 gene markers
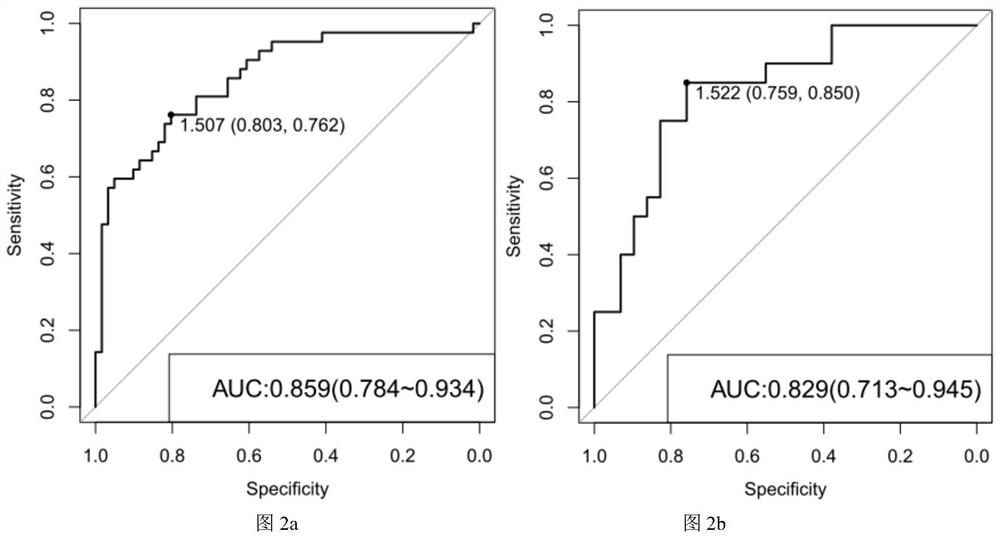
[0103]In order to verify the findings in Example 2, the inventors further analyzed the abundance of the 44 genes shown in Table 1 in the feces samples of 20 healthy people and 29 Graves' disease patients in the verification group, and based on the verification situation. The 44 genes were deleted, and the DNA extraction, sequencing and gene abundance analysis of the verified population were performed with reference to Example 1.
[0104] The verification results are as follows: Among the 41 genes enriched in the healthy population, 40 genes were verified with high quality in the verification set (p value <0.02), and the p values of the gene markers enriched in the healthy population in the verification group were The values are shown in Table 2.
[0105] Table 2:
[0106]
[0107]
[0108] For the above-mentioned enrichment in Graves' disease patients, all three genes were verified with high quality in the validati...
Embodiment 3
[0202] The detection of embodiment 3 individual state
[0203] In this example, the inventors used 11 stool samples to detect the individual status of the sample source.
[0204] Determine the gene abundances of GA20_GI_0029955, GA61_GI_0091802, and C02_GI_0042821 shown in Table 3 in each stool sample with reference to the method in Example 2, and determine whether the abundances of these three genes in each sample fall within the respective levels in the disease control group or the healthy control group. The 95% confidence interval of the abundance, it is determined that the abundance of these three genes all fall into the corresponding interval of the disease group, and the individual corresponding to the sample is a Graves' disease patient, and it is determined that the abundance of the three genes all fall into the The status of the individual corresponding to the sample in the corresponding interval of the healthy group is non-Graves' disease patient.
[0205] The resul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com