Exosome markers for diagnosing lymph node metastasis of invasive breast cancer and application of exosome markers
A technology for lymph node metastasis and infiltration, applied in the field of detection and diagnosis, to achieve high sensitivity, good specificity, and avoid excessive tissue biopsy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] The process of screening exosome markers for diagnosing lymph node metastasis of invasive breast cancer will be described in detail below.
[0056] (1) Extraction and identification of exosomes
[0057] 1. Collection of serum samples
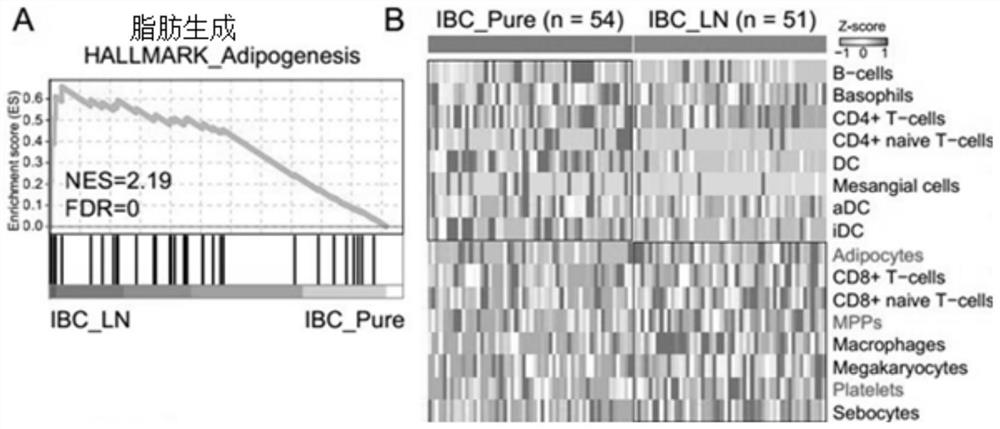
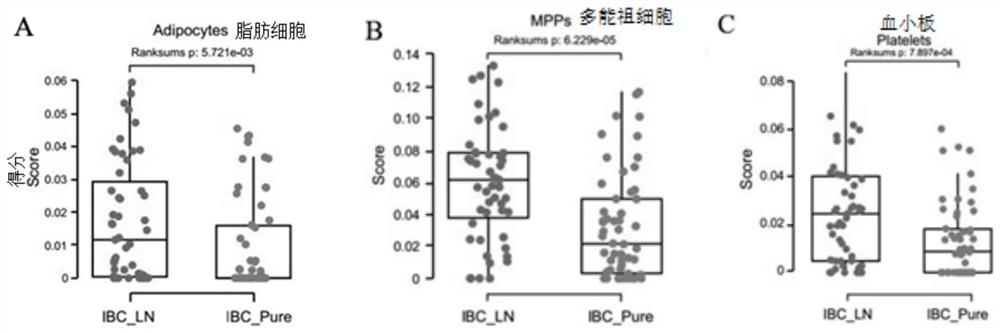
[0058] The serum samples of invasive breast cancer with lymph node metastasis (ibc_ln group) were from 51 patients diagnosed by Shanghai First People's Hospital Affiliated to Medical College of Shanghai Jiao Tong University; Serum samples of in situ invasive breast cancer (ibc_pure group) were also obtained from 54 patients in Shanghai First People's Hospital Affiliated to Medical College of Shanghai Jiao Tong University. These serum samples were placed in the refrigerator at - 80 ℃ for detection.
[0059] 2. Extraction of exosomes
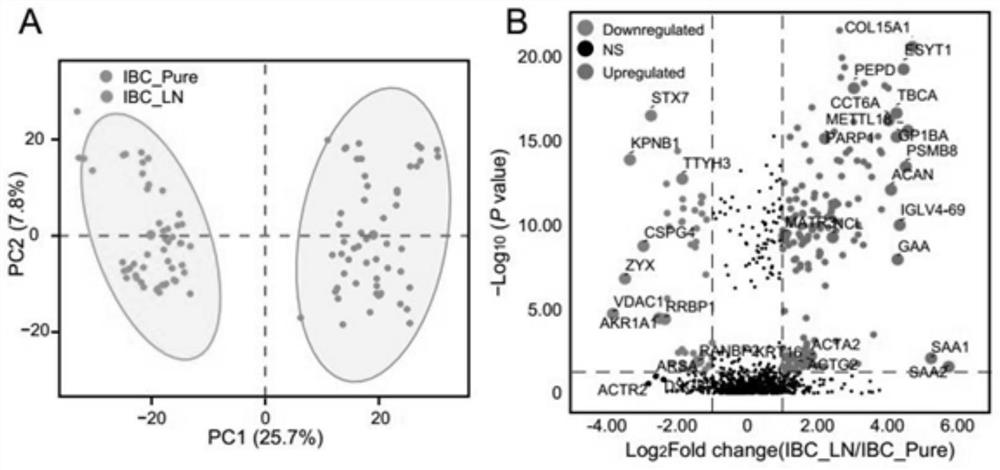
[0060] 1 ml serum was thawed on ice and centrifuged at 4 ℃ and 3000 RCF for 10 min; Take the supernatant, centrifuge with 10000g for 20min, dilute the supernatant with 25ml PBS and pass through 0.22 μ M centrifug...
Embodiment 2
[0084] Example 2 Establishment and verification of classifier
[0085] Based on the above research, the invention adopts the random forest classification method to build a molecular classifier to distinguish the lymph node metastasis of invasive breast cancer and in situ invasive breast cancer, and imports the above 30 significantly differentially expressed SEVs protein data into the random forest classification model to determine the SVS protein proton set that accurately distinguishes the lymph nodes of invasive breast cancer from the pure compartments of invasive breast cancer. In order to train and then test the model, the SEVs samples of the above two types of people are evenly divided, 70% of the samples are used as the training set, and the remaining 30% are used as the independent test set.
[0086] Results such as Figure 4 As shown in the figure below: the combination of 12 sev proteins (PEPD, NCL, PARP1, ACTA2, ACTG2, TBCA, matr3, krt16, CCT6A, ttyh3, KPNB1 and RanBP2) w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com