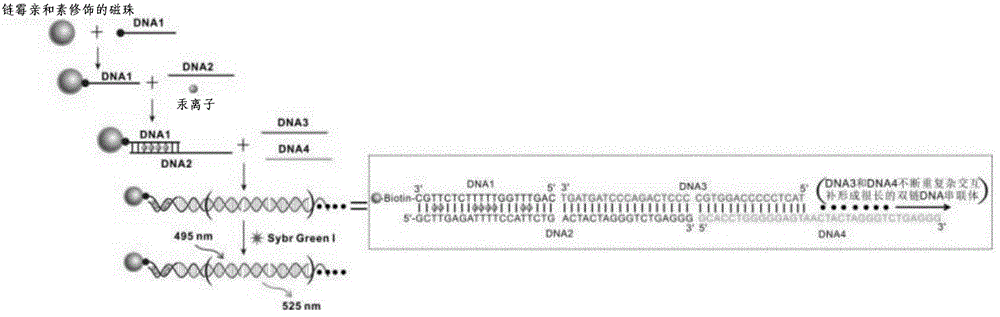
Mercury ion detection method based on nucleic acid probe head-to-tail complementation strategy, and mercury ion detection kit based on nucleic acid probe head-to-tail complementary strategy
A detection kit and detection method technology, which can be used in material excitation analysis, fluorescence/phosphorescence, etc., can solve the problems of low detection sensitivity, troublesome operation, increase experimental cost, etc., and achieve the effects of high sensitivity, simple operation and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] A detection kit for mercury ions based on the nucleic acid probe head-to-tail complementary strategy, including the following components:
[0041] (1) Streptavidin-modified magnetic beads;
[0042] (2) Biotin-modified DNA1, the sequence is as follows:
[0043] 5'-CAGTTTGGTTTTCTCTTGC-Biotin-3' (SEQ ID NO. 1)
[0044] (3) DNA2, the sequence is as follows:
[0045] 5'-GCTTGAGATTTTTCCATTCTGACTACTAGGGTCTGAGGG-3' (SEQ ID NO. 2)
[0046] (4) DNA3 and DNA4, the sequences are as follows:
[0047] DNA3: 5'-TACTCCCCCAGGTGCCCCTCAGACCCTAGTAGT-3' (SEQ ID NO. 3);
[0048] DNA4: 5'-GCACCTGGGGGAGTAACTACTAGGGTCTGAGGG-3' (SEQ ID NO. 4).
[0049] The 5' end of DNA3 is complementary to the 5' end of DNA4, the 3' end of DNA3 is complementary to the 3' end of DNA4, and the 3' end of DNA3 is complementary to the 3' end of DNA2;
[0050] (5) 20 mM Tris-acetate buffer (pH 7.5, containing 50 mM sodium acetate).
[0051] (6) Sybr Green I solution.
[0052] The working principle of this kit...
Embodiment 2
[0058] The method for detecting mercury ions based on the head-to-tail complementary strategy of nucleic acid probes is carried out according to the following steps:
[0059](1) Add 1 mM biotin-modified DNA1 to the streptavidin-modified magnetic bead solution, mix well, react at room temperature for 30 minutes, separate the magnetic beads, and remove excess DNA1.
[0060] (2) The magnetic bead-DNA1 mixture was resuspended with 20 mM Tris-acetate buffer (pH 7.5, containing 50 mM sodium acetate), and then 1 mM DNA2 was added, as well as Hg 2+ , mix well, react at room temperature for 30 minutes, separate with magnetic beads, and remove excess DNA2.
[0061] (3) The above mixture was resuspended with 20 mM Tris-acetic acid buffer (pH 7.5, containing 50 mM sodium acetate), then added 2 mM DNA3 and 2 mM DNA4, reacted at room temperature for 60 minutes, and separated with magnetic beads to remove excess DNA3 and DNA4.
[0062] (4) The above mixture was resuspended in 20 mM Tris-ace...
Embodiment 3
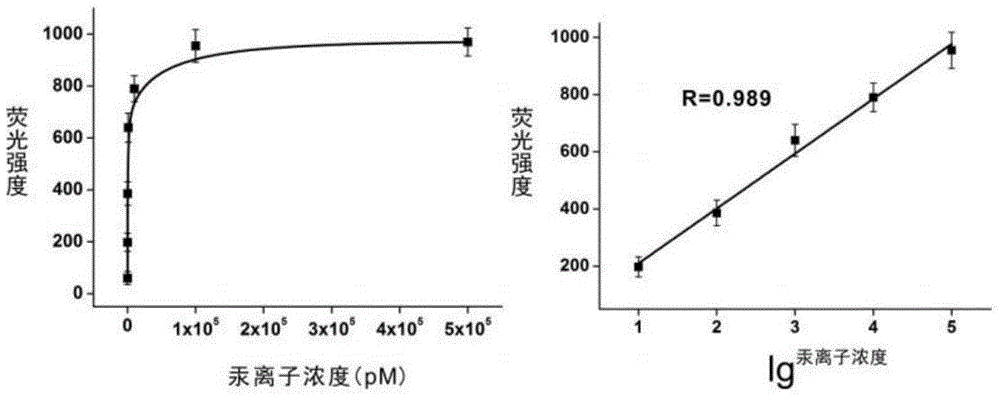
[0064] For different concentrations of Hg 2+ detection of:
[0065] Prepare Hg 2+ Standard solutions with concentrations of 10pM, 100 pM, 1 nM, 10 nM, 100 nM and 500 nM were stored at room temperature.
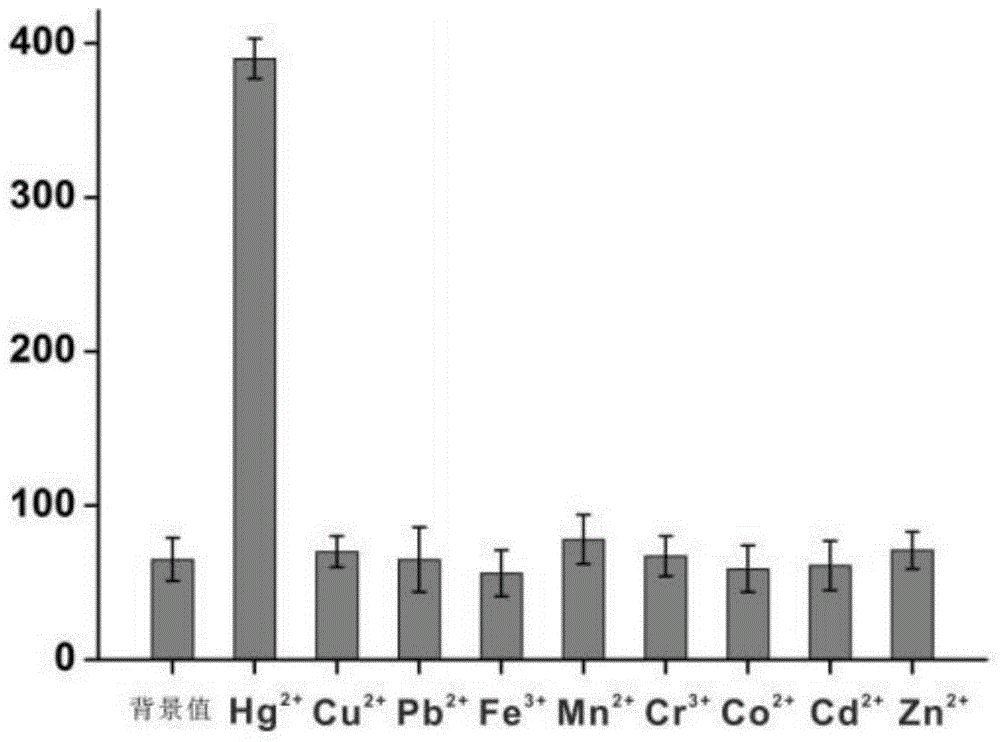
[0066] Different concentrations of Hg 2+ Solution is added in the reaction system described in embodiment 1 respectively, detects fluorescence intensity after sufficient reaction, as figure 2 As shown, with Hg 2+ As the concentration increases, the corresponding fluorescence intensity also increases, when Hg 2+ When the concentration exceeds 100 nM, it gradually reaches saturation. in Hg 2+ Logarithm of concentration (lg 汞离子浓度 ) is the abscissa, the fluorescence intensity is the ordinate, draw the standard curve, the two have a good linear relationship, the linear range is from 10pM to 100 nM, the linear equation is: F = 191.8 lg C + 18.4 (R=0.989), according to the standard of 3 times signal-to-noise ratio (3S / N), the detection limit is 2 pM.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com