GPU parallel computing molecular similarity method, device and system and medium
A parallel computing and similarity technology, used in computing, multi-programming devices, and analyzing two-dimensional or three-dimensional molecular structures, etc., can solve problems for a long time, and achieve the effect of improving computing speed.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0042] In order to make the object, technical solution and effect of the present invention more clear and definite, the present invention will be further described in detail below. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
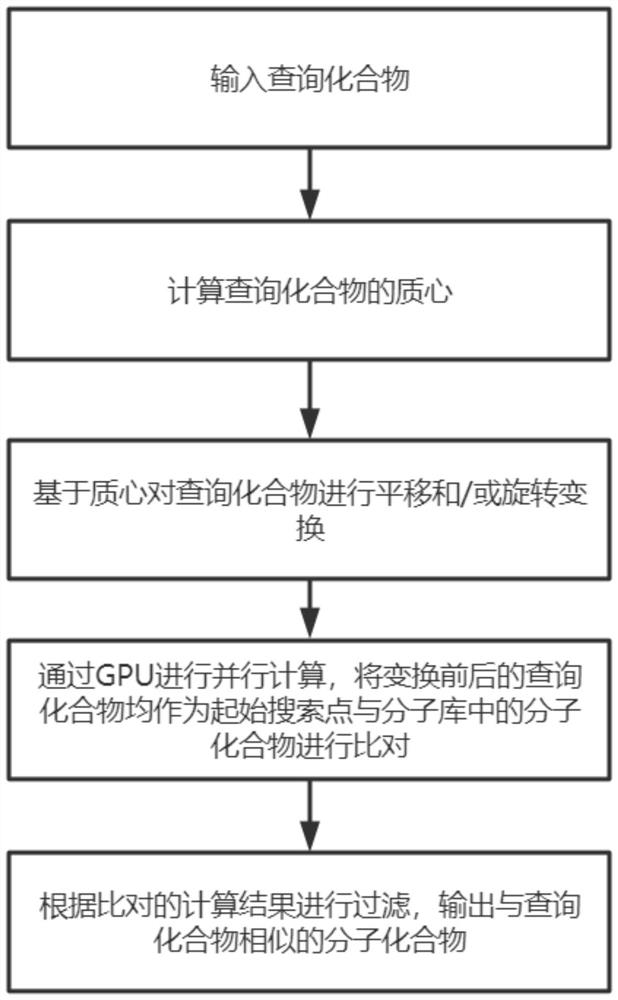
[0043] Such as Figure 1-2 Shown, a kind of GPU parallel computing molecular similarity method comprises the following steps:
[0044] S100, input query compound;
[0045] Extract from the database or custom build, input the compound to be queried, extract feature points based on the queried compound, and generate a search formula.
[0046] S200. Calculate the centroid of the query compound;
[0047] When calculating the 3D Gaussian superposition between the query compound and the compounds in the library, in order to ensure the accuracy of the calculation and improve the calculation speed, the query compound will be adjusted. In order to facil...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com