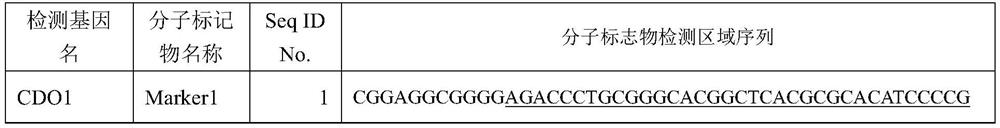Methylated molecular marker for detecting benign and malignant pulmonary nodules or combination and application thereof
A technology of molecular markers and methylation, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of insufficient molecular detection of tissue samples, avoid excessive diagnosis and treatment, reduce False positive rate and the effect of improving the detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
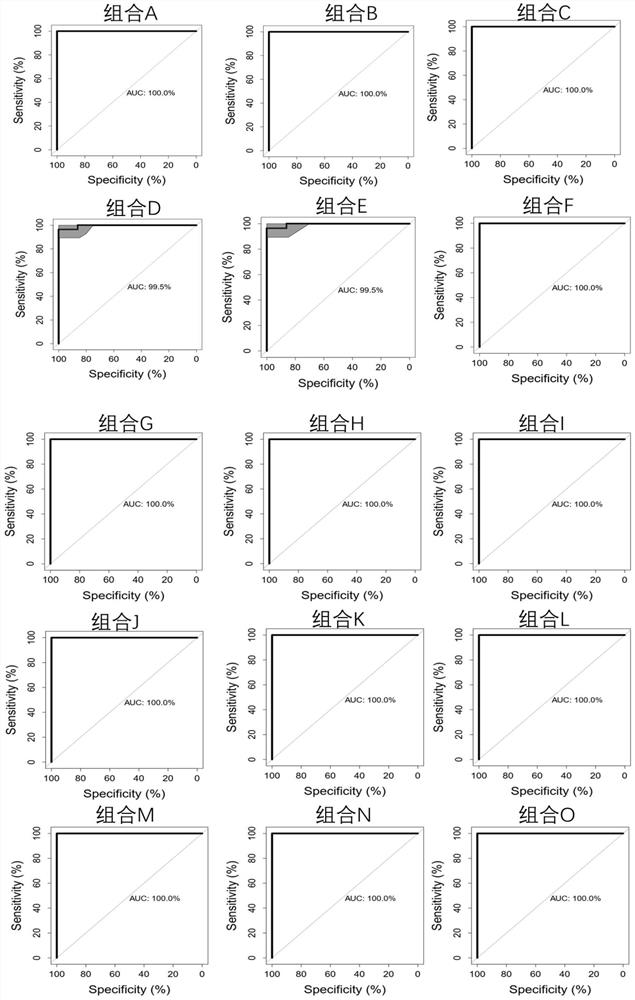
[0151] A kit for the detection of benign and malignant pulmonary nodules in respiratory samples, including the DNA of 19 methylated regions in 10 detection genes CDO1, DAPK, GHSR, HOXA11, HOXB4, LHX9, MIR196A1, PTGER4, SHOX2 and TBX15 The detection primers and probes of the methylation molecular markers Marker1 to Marker19, the detection region sequence and sequence number of each DNA methylation molecular marker are specifically shown in Table 1 (the underlined part of each region is the following embodiment of the present invention Marker of the corresponding sequence of the fragment amplified by the preferred primers used):
[0152] Table 1. Molecular marker detection region sequence
[0153]
[0154]
[0155]
[0156] The kit has designed three pairs of primers and three probes for each of the specific methylation sites in the 19 molecular markers Marker1 to Marker19 for the detection of benign and malignant pulmonary nodules in respiratory samples (probe fluoresc...
Embodiment 2
[0169] In this example, the kit described in Example 1 is used to detect the methylation levels of Marker1 to Marker19 in respiratory samples.
[0170] The method for detecting the methylation level of the DNA methylation molecular marker comprises the following steps:
[0171] 1. Extraction of gDNA from respiratory samples:
[0172] 1) Extraction of gDNA from paraffin section samples of lung tissue: the specific operation steps of gDNA extraction from paraffin tissue were carried out according to the instructions of Qiagen's ALLPrep DNA / RNA FFPE Kit;
[0173] 2) Extraction of gDNA from the respiratory fluid sample: First, the respiratory fluid sample was centrifuged at a low speed at 4°C, 5000g, for 5min; the supernatant was removed, and the precipitate was collected; Blood&Tissue Kit instructions were used to extract gDNA.
[0174] 2. Perform sulfite conversion on the extracted gDNA
[0175] The extracted gDNA is subjected to bisulfite conversion, and 50-100ng of gDNA is...
Embodiment 3
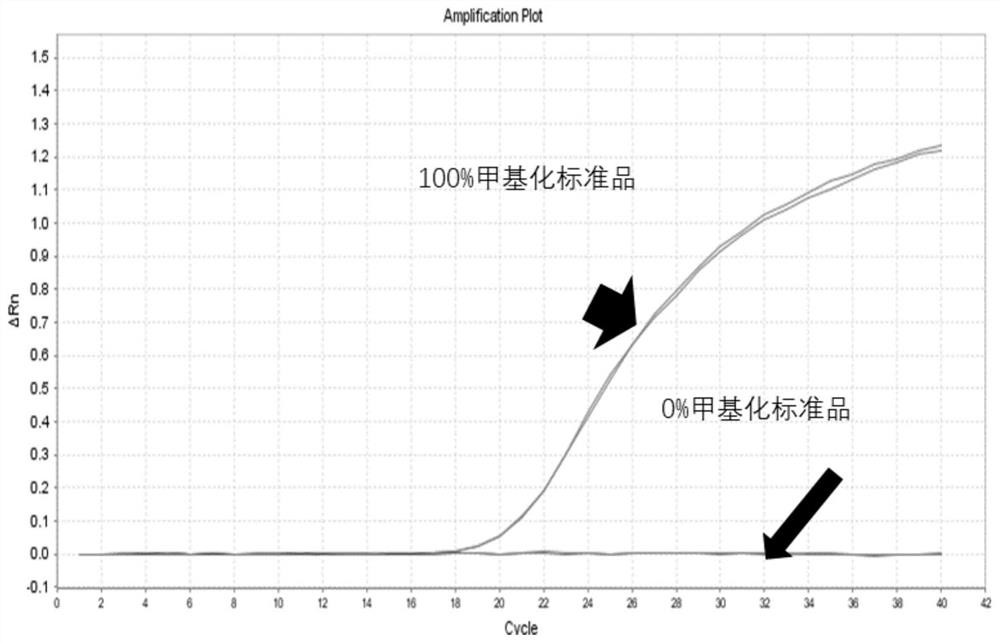
[0191] This embodiment provides a detection method for molecular markers in standard products, and the detection steps are as follows:
[0192] 1. Standard product preparation
[0193] 1) Preparation of 0% methylation standard:
[0194] use Single Cell Kit (Qiagen, Cat#150343) and Mung Bean Nuclease (NEB, Cat#M0250L) were used to process NA12878 DNA to prepare 0% methylation standard;
[0195] 2) Preparation of 100% methylation standard:
[0196] The prepared 0% methylation standard was treated with CpG Methyltransferase (M.SssI) to obtain a 100% methylation standard.
[0197] 2. Preparation of standard products with different methylation ratios:
[0198] Mix 0% and 100% methylation standards according to the desired methylation ratio gradient to obtain 0.2%, 0.4%, and 1% methylation ratio standards.
[0199] 3. Perform bisulfite conversion on the standard DNA with different methylation ratios: the steps are as in Example 2, and the conversion input amount is 50-100 ng, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com