Molecular marker related to degradation and self-capping of watermelon tendrils and application of molecular marker
A molecular marker, self-capping technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as unclear regulation mechanism of self-capping traits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
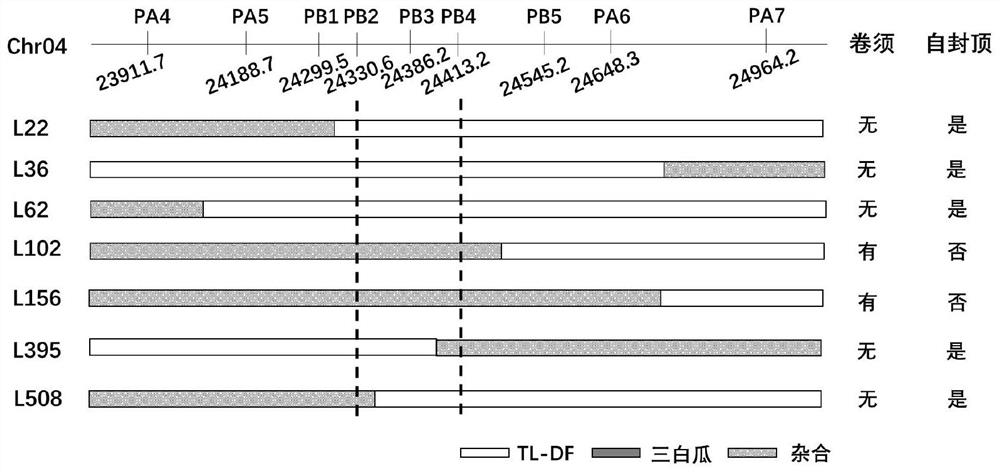
[0029] Example 1. Acquisition of InDel marker ID_PB3 co-isolated from watermelon tendril degeneration and self-capping gene
[0030] 1. Materials and group construction
[0031] F1 obtained by crossing TL-DF (watermelon material with degenerated tendrils and self-capping) as the female parent, with Sanbaigua (watermelon material with normal tendrils growing indefinitely) as the male parent, and using the above-mentioned TL-DF female parent and Sanbaigua as the male parent. Generation, the F1 generation backcrossed with TL-DF to obtain the BC1P1 generation population, while the F1 generation self-crossed one generation to obtain the F2 generation population.
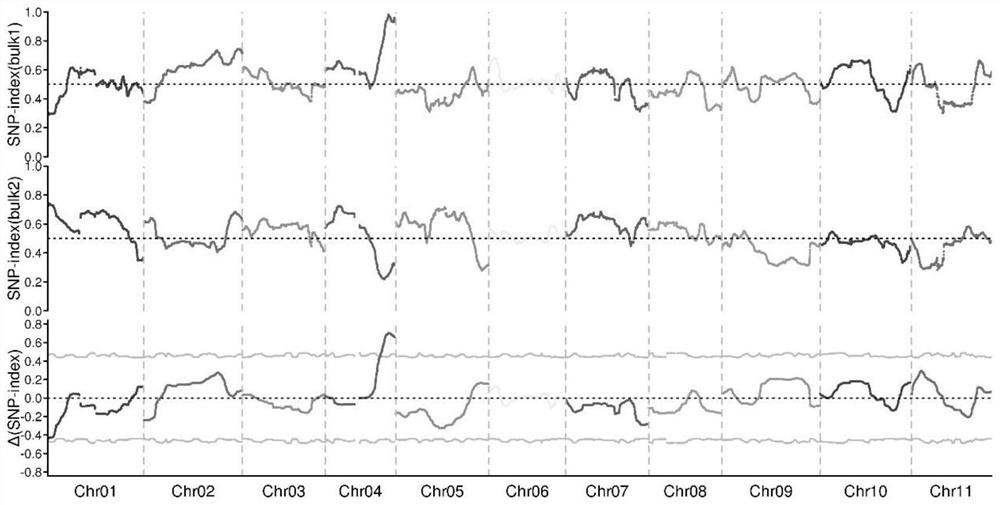
[0032] 2. Preliminary localization of tendril degeneration and self-capping genes
[0033] In the spring of 2021, the parental TL-DF, Sanbaigua, F1 generation plants, 113 F2 individual plants, and 62 BC1 individual plants were planted in Wuhan, and the tendril degradation and self-capping traits were identified on the pa...
Embodiment 2
[0065] Example 2. Identification of watermelon tendrils and self-capping traits
[0066] In order to further verify the linkage relationship between the ID_PB3 marker in Example 1 and the watermelon flesh color gene, 50 natural populations were used for verification. Specific steps are as follows:
[0067] 1. Materials to be tested
[0068] The test materials are 50 watermelon natural resource groups, all of which are normal tendrils and infinite inflorescence growth type (see Table 4 for details); each test material in Table 4 is preserved by the melon research group of the Economic Crops Research Institute of Hubei Academy of Agricultural Sciences. Germplasm resources.
[0069] Table 4 Identification results of individual plant materials and tendrils and inflorescence growth types of natural populations composed of 50 watermelon germplasm resources
[0070] (+ / + in marking detection means tendrils degenerate and self-capping, + / - and - / - means tendrils grow normally and i...
Embodiment 3
[0083] Example 3. Molecular-assisted breeding of watermelon tendrils and inflorescence growth traits
[0084] Molecular marker-assisted breeding using the molecular marker ID_PB3 obtained in Example 1 using tendril degeneration and inflorescence self-capping watermelon material, including the following steps: DNA extraction: using tendril degeneration and self-capping watermelon as donor parents, other tendrils grow normally and indefinitely Watermelon is the recipient parent, and the substitution line and its derivative lines obtained by hybridization and multi-generation backcross are used to extract the genomic DNA of the isolated population by CTAB (Paterson et al. 1993) method at the seedling stage;
[0085] Use the molecular marker ID_PB3 to detect the genotype of the individual plants of the above population, and select the individual plants whose ID_PB3 marker is heterozygous (+ / -) at the 67th position in each generation for the next round of backcrossing; after 6-8 con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com