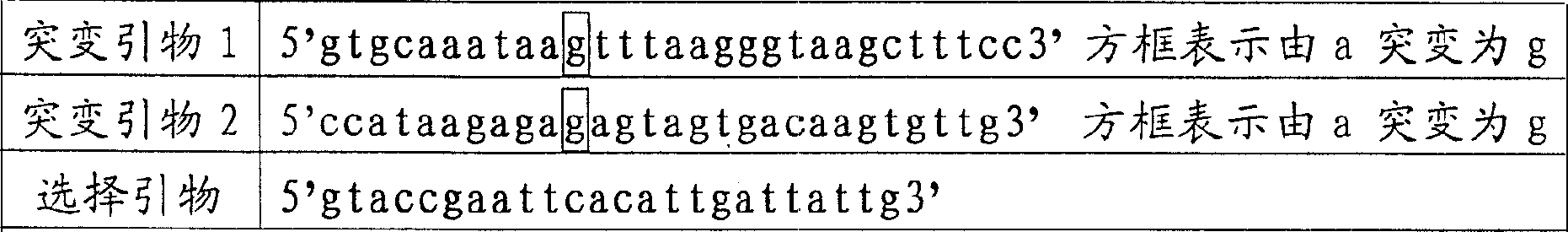Method of point mutation without need of chain reaction of polymerase and the kit
A technology of chain reaction and polymerase, which is applied in the direction of mutant preparation, biochemical equipment and methods, and microbial measurement/inspection. It can solve the problems of time-consuming operation and cumbersome steps, and achieve the effect of low price and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The molar ratio of primer (phosphorylated at the 5' end) to template is 100:1
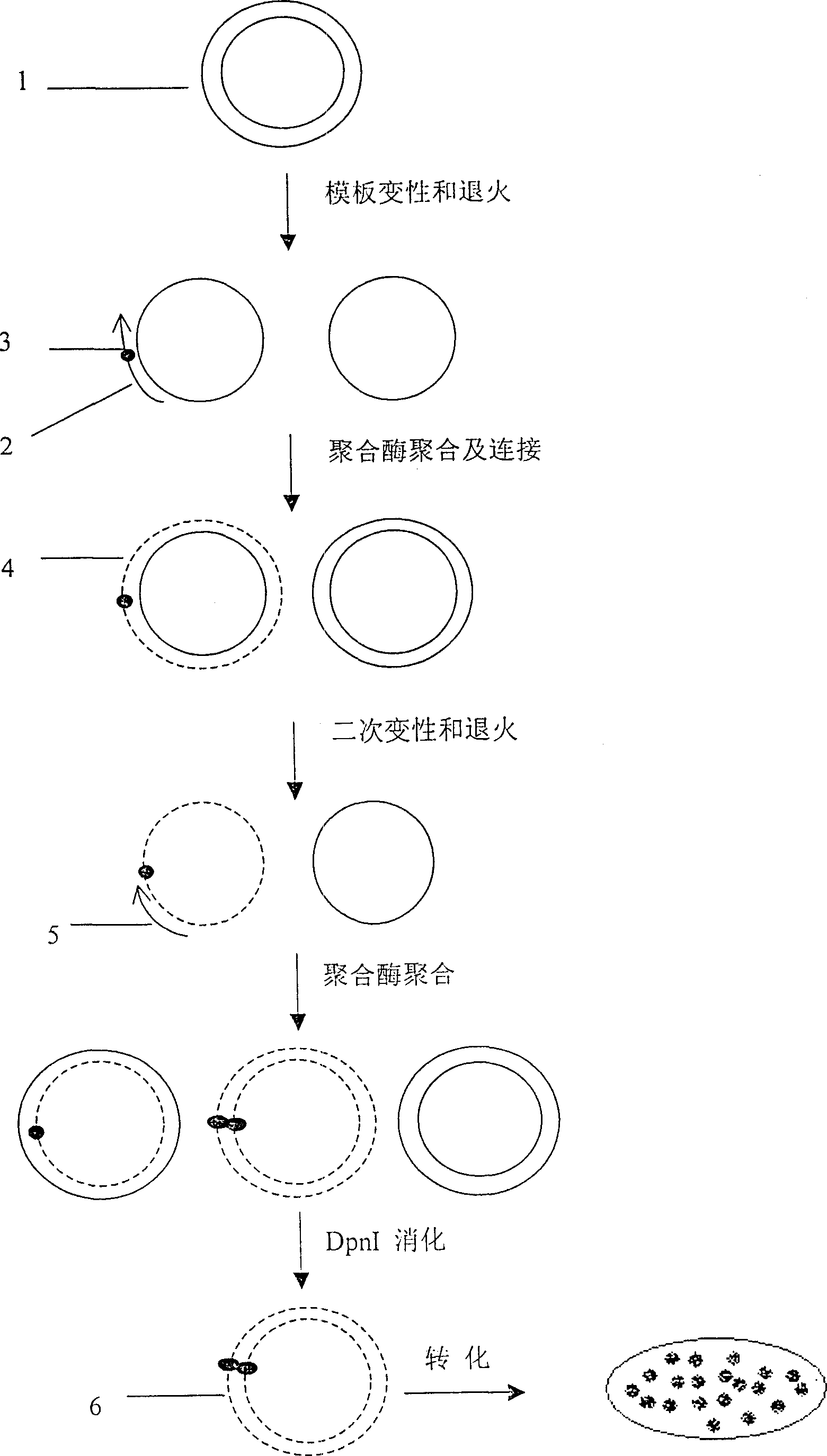
[0042] 1. The first step is template denaturation and primer annealing to template
[0043] Template 500ng
[0044] Mutation primer 1 130ng
[0045] Annealing buffer 1 2μl
[0046] H 2 O 20-X
[0047] Reaction conditions: the above mixture was reacted at 100° C. for 5 minutes; rapidly ice bathed for 5 minutes; room temperature for 30 minutes to obtain an annealed mixture.
[0048] X means except H 2 The sum of the volumes of other solutions other than O (the following explanations are the same for X in each embodiment).
[0049] 2. Mutant strand synthesis and nick ligation
[0050] Annealing mixture 20μl
[0051] 10× synthesis buffer 3μl
[0052] T4 DNA polymerase 1μl
[0053] T4 DNA ligase 1 μl
[0054] 10mM dNTPs 1.5μl
[0055] H 2 O 3.5 μl
[0056] Ice bath for 5 minutes, room temperature for 5 minutes, 37°C, water bath for 2 hours.
[0057] 3. The second step of dena...
Embodiment 2
[0076] The molar ratio of the two point mutation primers (phosphorylated at the 5' end) to the template is 200:1
[0077] 1. The first step is template denaturation and primer annealing to template
[0078] Template 500ng
[0079] Mutation Primer 1 260ng
[0080] Mutation Primer 2 260ng
[0081] Annealing buffer 2 2 μl
[0082] H 2 O 20-X
[0083] 100°C for 5 minutes; rapid ice bath for 5 minutes; room temperature for 30 minutes to obtain an annealing mixture.
[0084] Step 2, 3, 4, 5, 6 and 7 are the same as in Example 1.
Embodiment 3
[0086] The molar ratio of primer (phosphorylated at the 5' end) to template is 100:1
[0087] 1. The first step template denaturation and primer annealing template
[0088] Template 500ng
[0089] Mutation primer 1 130ng
[0090] Annealing buffer 1 2μl
[0091] H 2 O 20-X
[0092] 100°C for 5 minutes; rapid ice bath for 5 minutes; room temperature for 30 minutes to obtain an annealing mixture.
[0093] 2. DNA Polymerization and Nick Ligation
[0094] Annealing mixture 20μl
[0095] 10× synthesis buffer 3 μl
[0096] T7 DNA polymerase 1μl
[0097] T4 DNA ligase 1 μl
[0098] 10mM dNTPs 1.5μl
[0099] H2 O 3.5 μl
[0100] 5 minutes in ice bath, 5 minutes at room temperature, 37°C, 30 minutes in water bath.
[0101] 3. The second step of denaturation and annealing
[0102] Add 100ng of selection primers, 100°C for 5 minutes; quickly ice bath for 5 minutes; room temperature for 30 minutes, to obtain a denaturation mixture.
[0103] 4. The second step of aggrega...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com