High-sensitivity homogeneous DNA hibridization fluorescence detection method
A fluorescence measurement, high-sensitivity technology, used in fluorescence/phosphorescence, material excitation analysis, etc., can solve the problem of low sensitivity, and achieve the effect of high sensitivity and wide application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
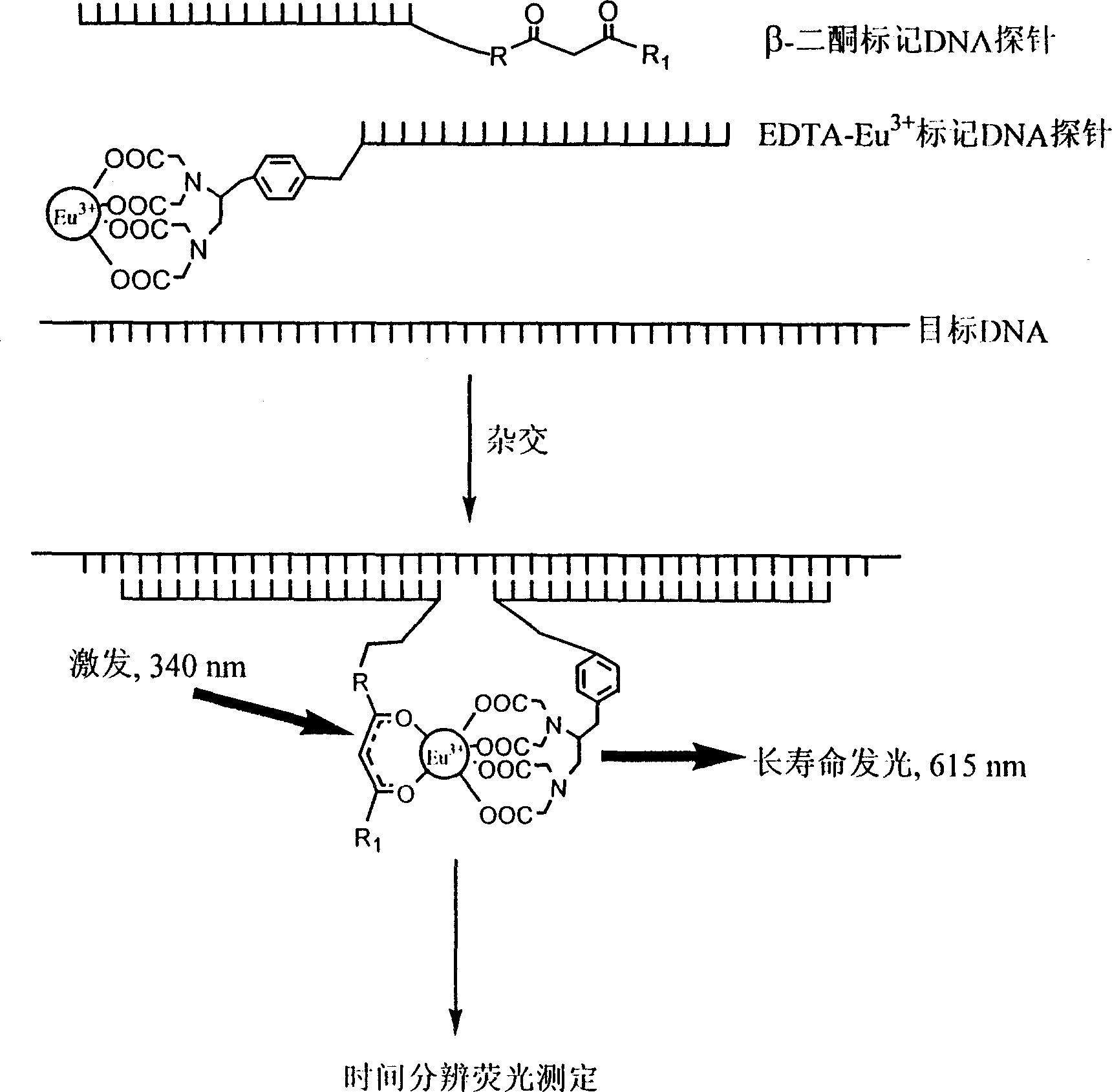
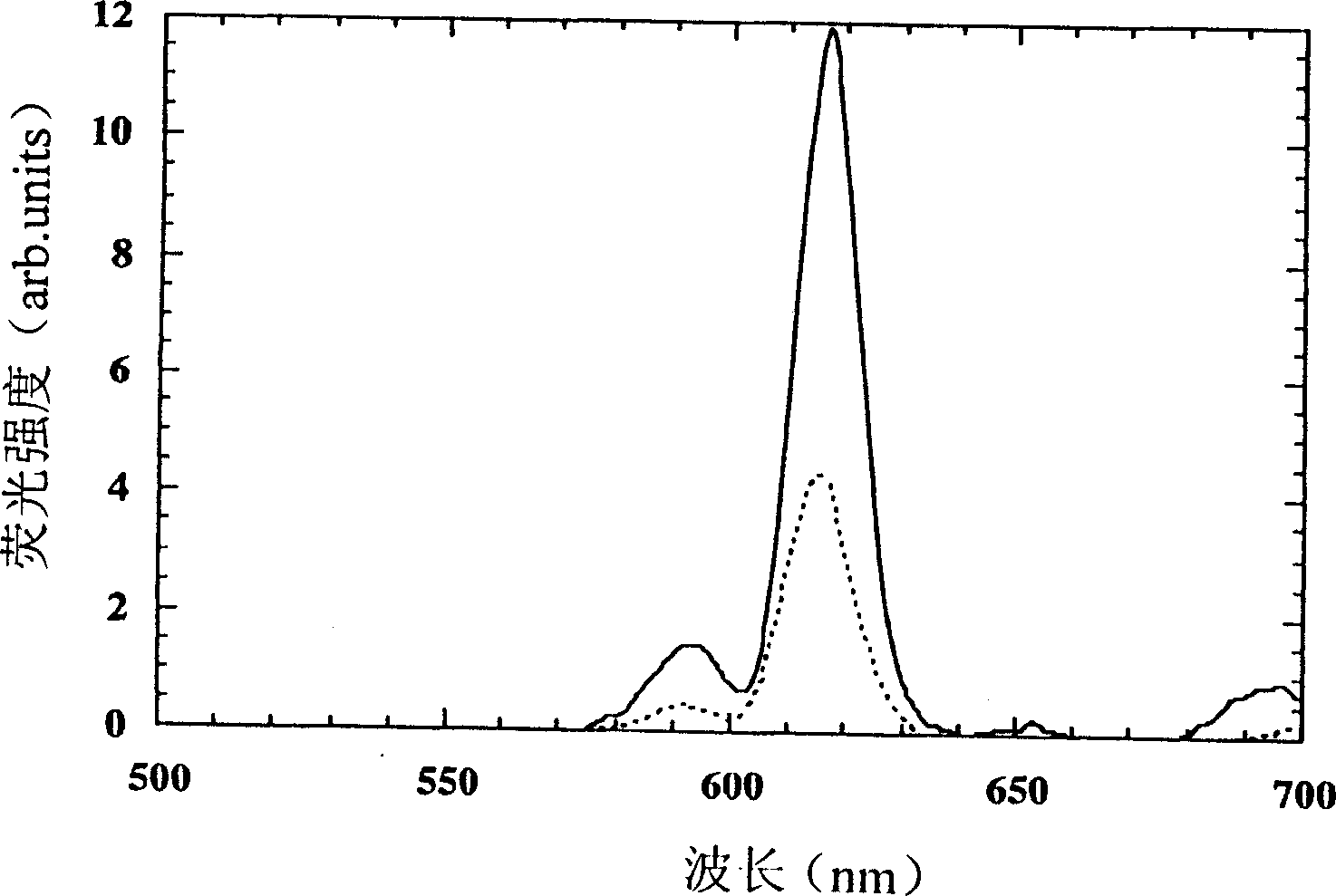
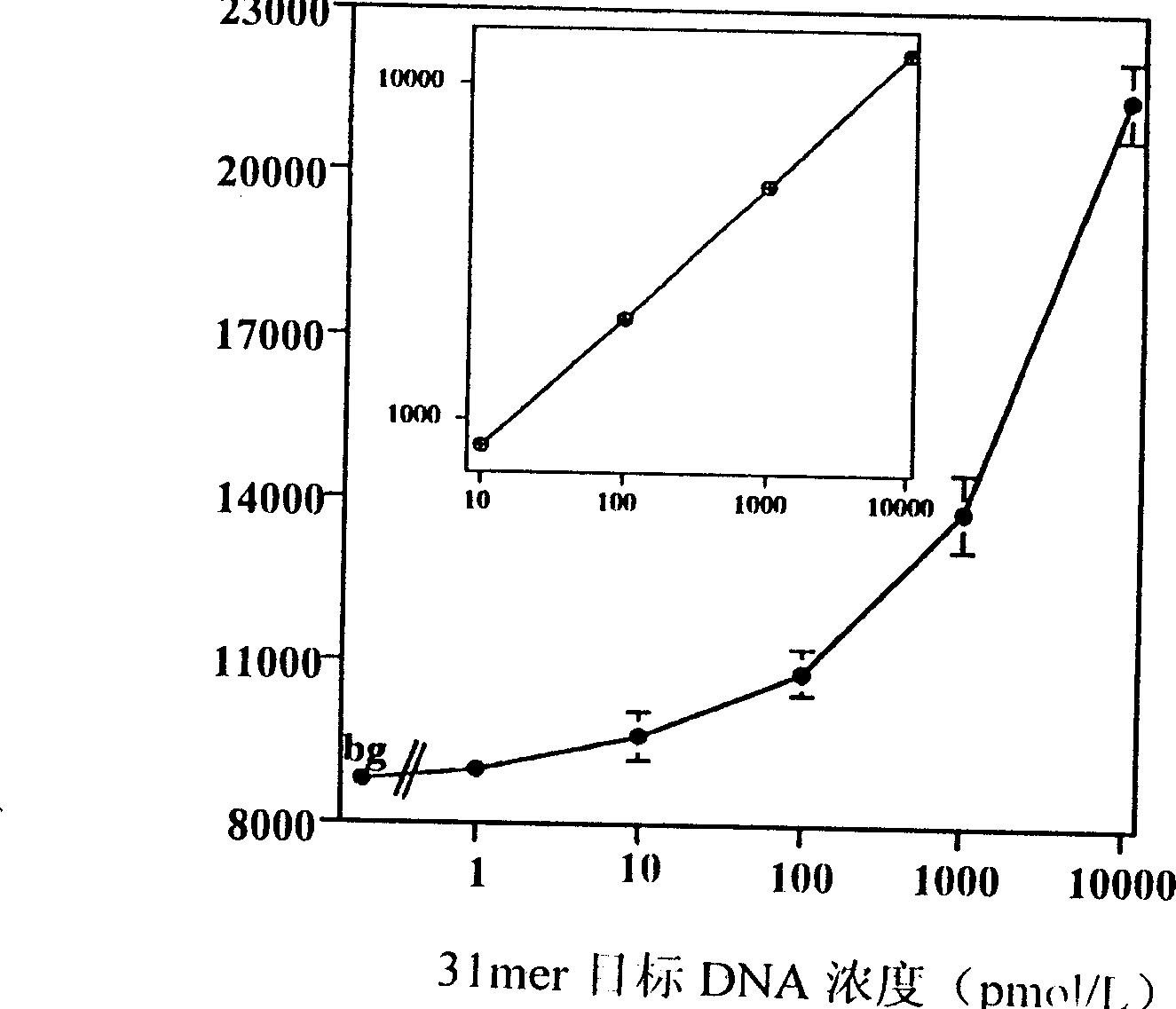
[0032] A DNA probe labeled at the 5'-end (or 3'-end) with a europium complex of EDTA and a DNA probe at the 3'-end (or 5'-end) labeled with a bidentate β-diketone DNA probes are used in DNA hybridization reactions. When the two labeled DNA probes hybridize to the target DNA, EDTA-Eu 3+ Close to bidentate β-diketone to form EDTA-Eu 3+ -β-diketone strongly fluorescent ternary fluorescent complex, the concentration of target DNA can be measured by measuring the fluorescence intensity of the formed ternary fluorescent complex by time-resolved fluorescence assay.
[0033] 1. Synthesis of bidentate β-diketone labels for labeling DNA probes
[0034] This example uses a novel bidentate β-diketone containing a chlorosulfonyl group, 5-(4"-chlorosulfonyl-1',1"-biphenyl-4'-yl)-1,1,1, 2,2-pentafluoro-3,5-pentanedione (CDPP) was used to label DNA probes. The synthesis reaction of CDPP is carried out in two steps, and its reaction principle is shown in the following reaction equation, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com