Fluorescent enzyme assay methods and compositions
A technology of composition and fluorescent part, which is applied in the field of luciferase analysis and composition, and can solve the problems of inconvenient and expensive analysis operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0246] a. Protein Kinase Substrate (Compound 1)
[0247] An exemplary enzyme substrate for the detection of protein kinase A, palmitoyl-FAM-S (OPO 3 2- )LRRRRFSK(Ac)G-amide. Use standard FastMoc TM Chemical method Construction of peptide Fmoc-L(R(Pmc)) via solid-phase peptide synthesis on 625mg of Fmoc-PAL-PEG-PS resin at 0.16mmol / g 4 FS(tBu)K(Mtt)G, the resin is a solid support for generating carboxyamide peptides. A portion of the final protected peptide-resin (20 mg, 2 μmol peptide) was transferred to a 1.5 ml Eppendorf tube and treated with 1 mL of 5% trifluoroacetic acid (TFA) in dichloromethane (DCM) to generate the characteristic yellow trityl base color. The resin was precipitated by adding 0.1 mL methanol and washing (3 x 1 mL dimethylformamide, DMF). Diisopropylethylamine (50 μL) and capping solution (1 mL of acetic anhydride solution (0.5 M) and hydroxybenzotriazole (0.015 M) in N-methylpyrrolidone (NMP)) were added to the re...
Embodiment 2
[0251] Phosphatase detection
[0252] a. Phosphatase substrate (compound 2)
[0253] An exemplary dye-labeled peptide is described below, compound 2, palmitoyl-LRRRFS (OPO 3 2- ) Synthesis of K(5-FAM)G-amide. Use standard FastMoc TM Chemical method Construction of peptide Fmoc-LR(Pmc) via solid-phase peptide synthesis at 0.16mmol / g on 625mg of Fmoc-PAL-PEG-PS resin 4 FS(OPO(OBzl)OH)K(Mtt)G, the resin is a solid support for generating carboxyamide peptides. Peptides with carboxyl termini were constructed with Fmoc-Gly-PEG-PS resin. A portion of the final protected peptide-resin (20 mg, 2 μmol peptide) was transferred to a 1.5 ml Eppendorf tube and treated with 1 mL of 5% trifluoroacetic acid (TFA) in dichloromethane (DCM) to generate the characteristic yellow triphenyl Methyl color. The resin was precipitated by adding 0.1 mL methanol and washing (3 x 1 mL dimethylformamide, DMF). 5-Carboxyfluorescein succinimidyl ester (5 mg, 10 μmol), diisopropylethylamine (30 μL,...
Embodiment 3
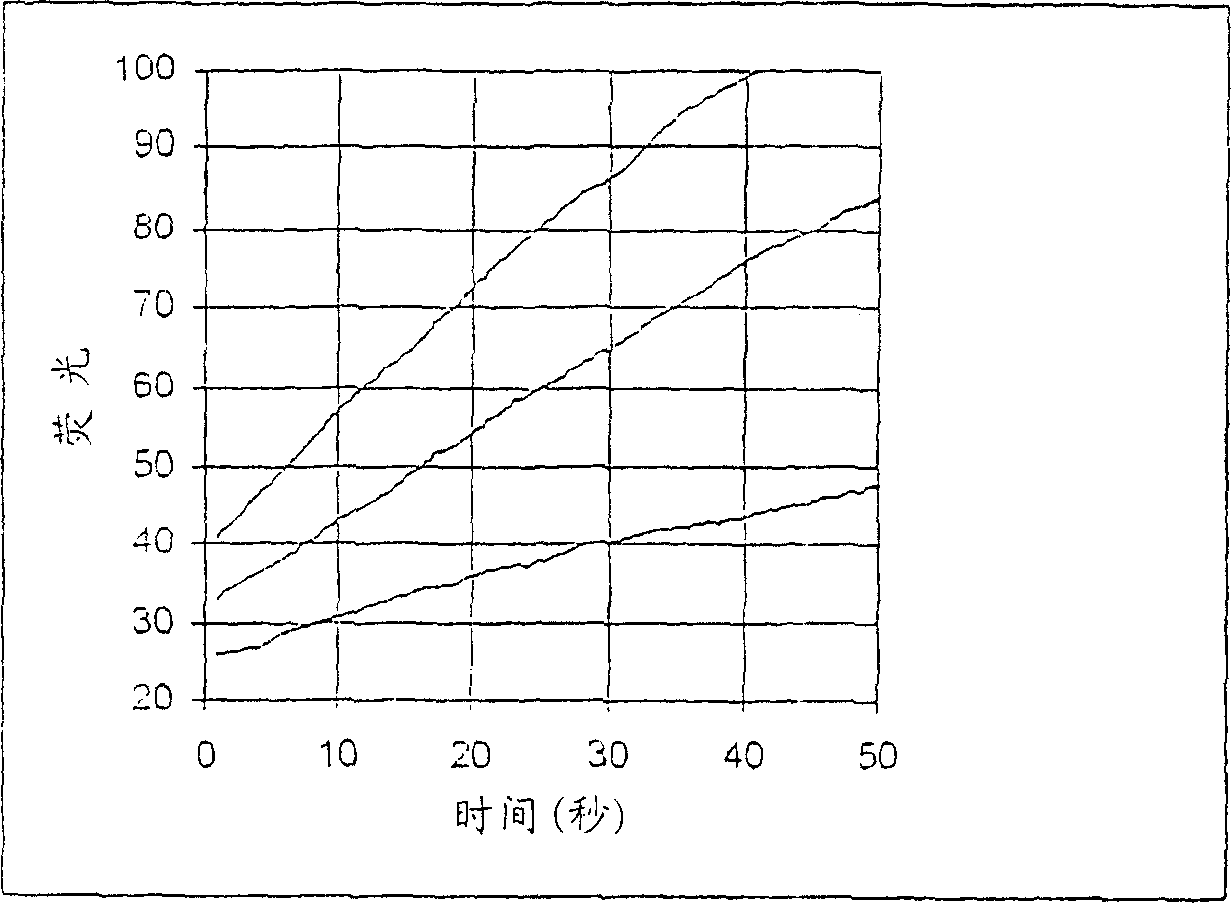
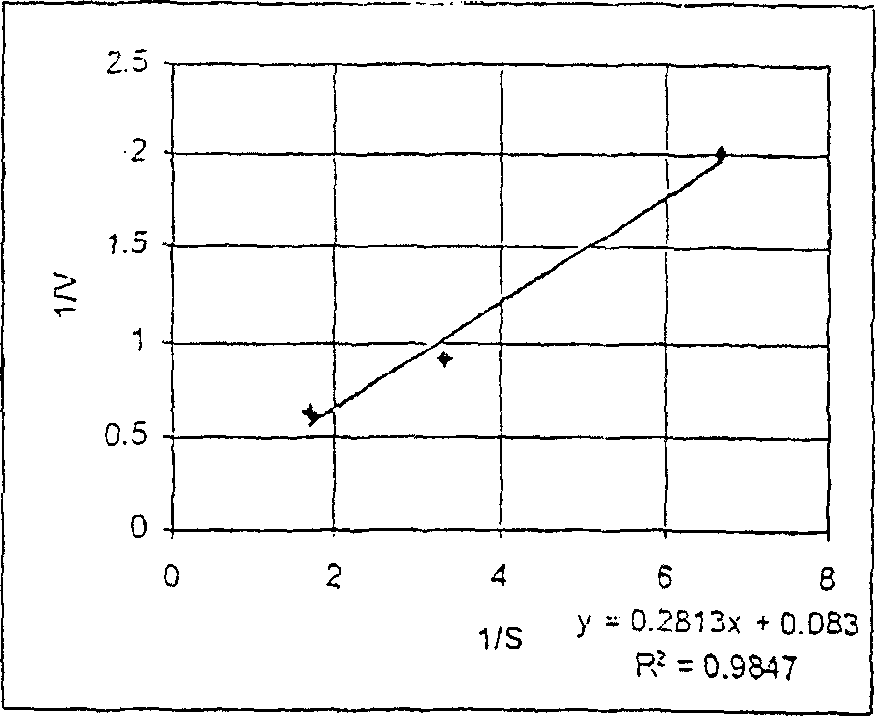
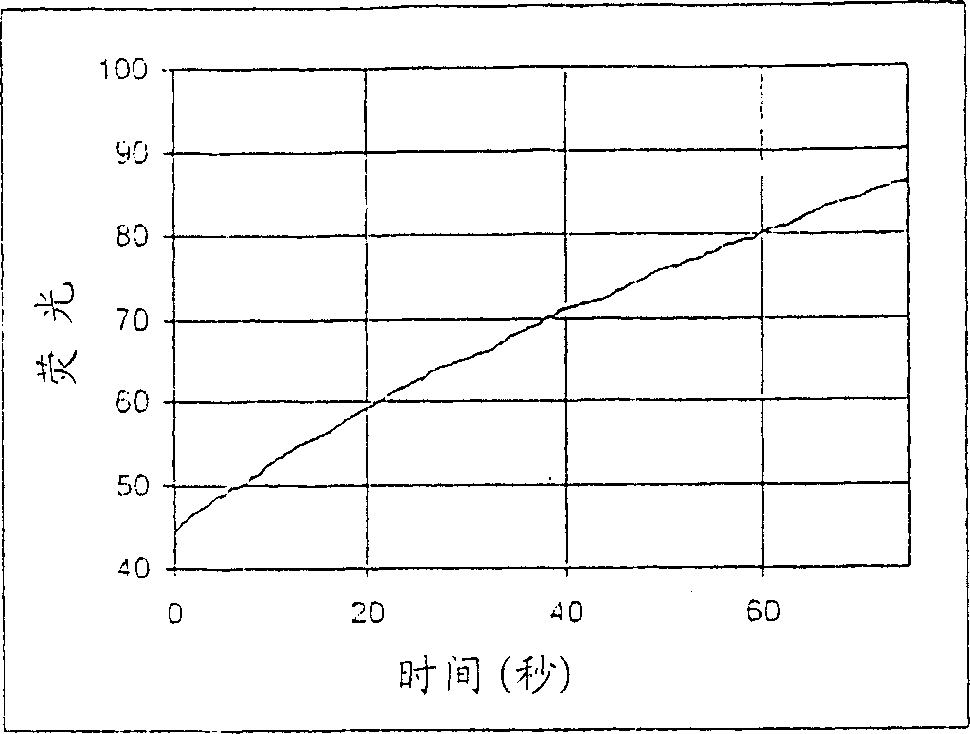
[0257] Fluorescence dynamic range
[0258] This example describes a study to determine the difference in fluorescence between phosphorylated and non-phosphorylated forms of a protein kinase A substrate. The goal is to determine the extent to which a compound provides an increase in fluorescent signal with a good signal-to-noise ratio after phosphorylation of the non-phosphorylated form.
[0259] a. Candidate substrates (compounds 3, 3P, 4, 4P, 5, 5P, 6 and 6P)
[0260] A series of compounds with alkylacyl groups of varying lengths (Scheme 3 above) were prepared in both phosphorylated and non-phosphorylated form, represented by the formula: X-Y(dye)LRRAS(OR)LG-NH 2 , where X is CH 3 (CH 2 ) x C(=O)-form fatty acid acyl group, x is 0, 7, 10 or 14, Y is α-aminomethylglycine, the dye is attached to 4,7-dichlorofluorescein at the 2-amino nitrogen atom of Y, and R is H or PO 3 2- .
[0261] For compounds 3-6, standard FastMoc TM Chemical method Construction of peptide F...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Extinction coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com