Nucleic acid and gene originating in novel hcv strain and replicon-replicating cell using the gene
一种复制子、核酸的技术,应用在基因工程、植物基因改良、生物化学设备和方法等方向,能够解决不适于建立自主感染并复制的细胞培养体系等问题,达到高概率复制效率的效果
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0129] Replication of replicon RNA
[0130] 1. Construction of expression vector
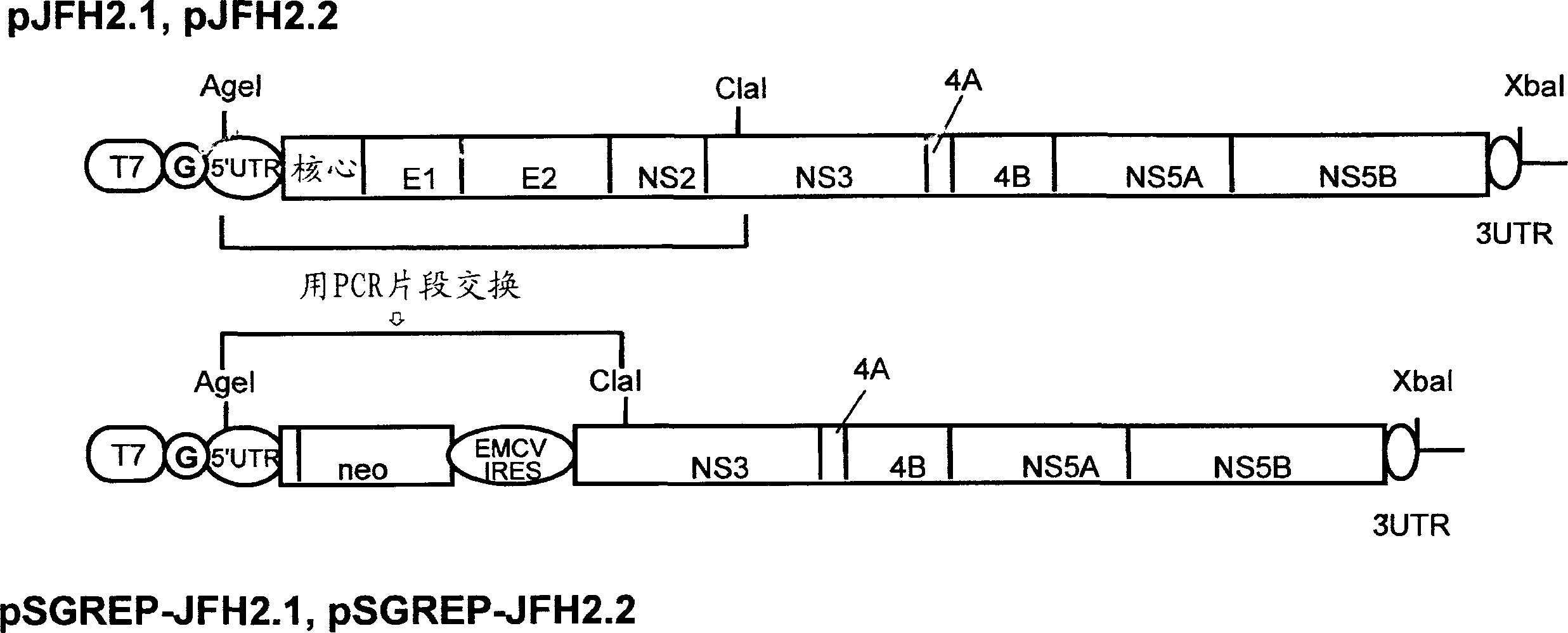
[0131] The DNA corresponding to the complete region of the genome of the hepatitis C virus JFH-2.1 strain and JFH-2.2 strain isolated from patients with fulminant hepatitis was obtained from the JFH-2.1 and JFH-2.2 clones containing the full-length genome cDNA of the virus strain, and the T7 RNA A synthetic DNA of a promoter sequence was attached to the 5' end of each clone, and then inserted into a plasmid (pUC19 plasmid). The plasmid DNAs thus constructed are hereinafter referred to as pJFH-2.1 and pJFH-2.2, respectively. It should be noted that the above JFH-2.1 and JFH-2.2 clones were prepared according to published reports (Lehmann et al., Science, (1999)).
[0132] The structures of the thus constructed plasmid DNAs pJFH-2.1 and pJFH-2.2 are shown in figure 1 line above. "T7" indicates the T7 RNA promoter, and "G" indicates the dGTP inserted upstream of the 5' end of pJFH-2.1 and pJFH-...
Embodiment 2
[0139] Establishment of replicon-replicating cell clones
[0140] For each synthetic replicon RNA rSGREP-JFH2.1 and rSGREP-JFH2.2 prepared in Example 1, 0.01 ng to 10 μg of replicon RNA was mixed with total cellular RNA extracted from Huh7 cells to adjust the concentration of total RNA The amount is 10 μg. The mixed RNA was then introduced into Huh7 cells by electroporation. The electroporated Huh7 cells were seeded in culture dishes and cultured for 16 hours to 24 hours, and then various concentrations of G418 (neomycin) were added to the dishes. Thereafter, the culture was continued while changing the medium twice a week. After 21 days of culture from the time of inoculation, surviving cells were stained with crystal violet. The result is as figure 2 Colony formation is confirmed as shown.
[0141] For cells transfected with rSGREP-JFH2.1 and rSGREP-JFH2.2 and showing colony formation, surviving cell colonies were further cloned from the culture dish after 21 days of c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com