Cloning, sequencing and expression of a gene encoding an eukaryotic amino acid racemase, and diagnostic, therapeutic, and vaccination applications of parasite and viral mitogens
a technology of eukaryotic amino acids and gene encoding genes, which is applied in the field of cloning, sequencing and expression of a gene encoding an eukaryotic amino acid racemase, and diagnostic, therapeutic, and vaccination applications of parasites and viral mitogens. it can solve the problems of i>trypanosoma cruzi , no effective treatment or vaccine against i>trypanosoma cruzi , the difficulty in the detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Mice and Parasites
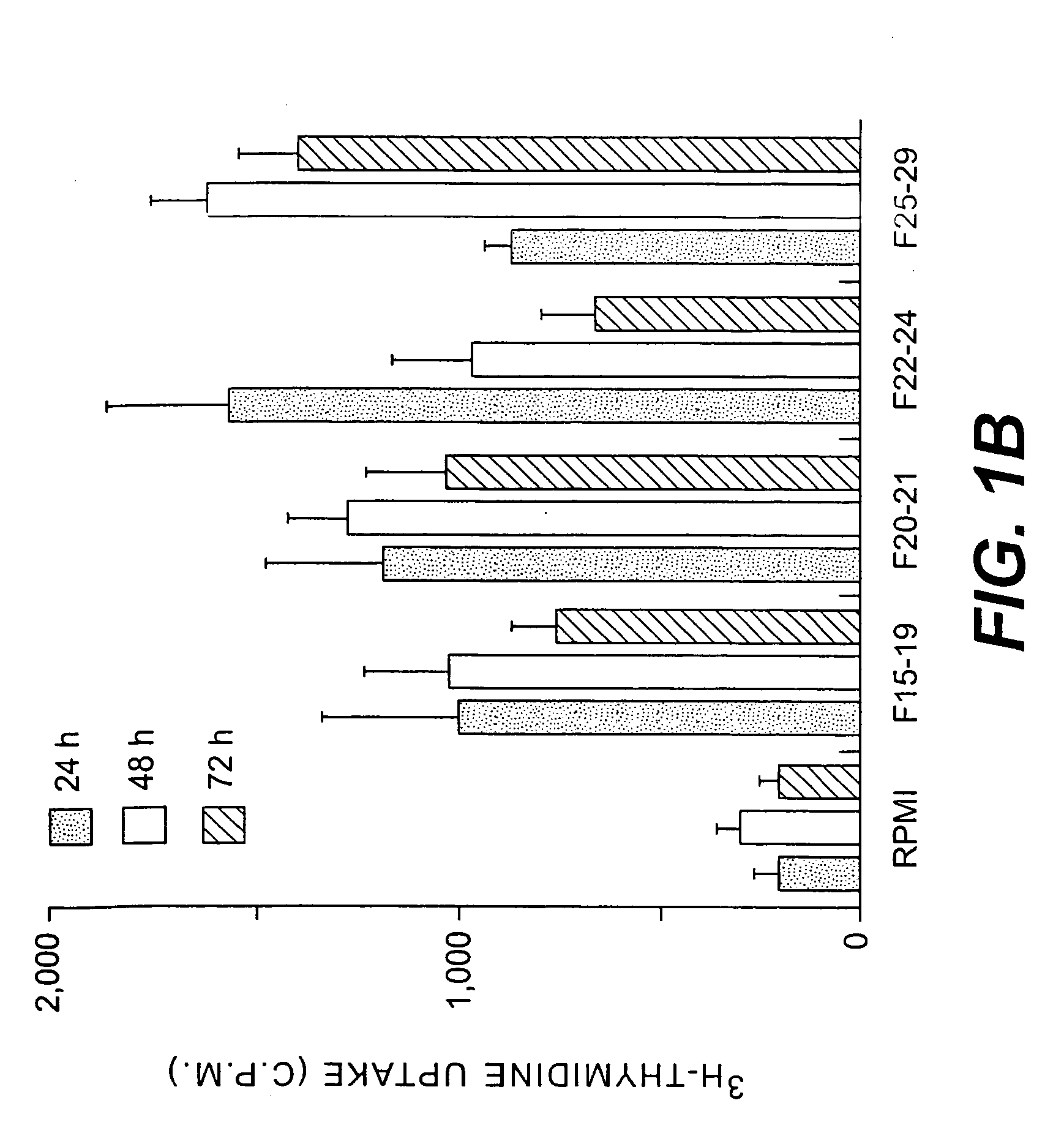
[0217]Trypanosoma cruzi clone CL Brener was used throughout this work. Epimastigotes were maintained by weekly passage in liver infusion tryptose medium. In vitro metacyclogenesis was performed in a protein free defined medium at 27° C., as previously described11. Male euthymic or athymic BALB / c mice 8 weeks of age were purchased from Charles River Laboratories (Saint Aubin les Elbeuf, France). Male C3H / HeI mice 8 weeks of age from our animal facilities were also used.
example 2
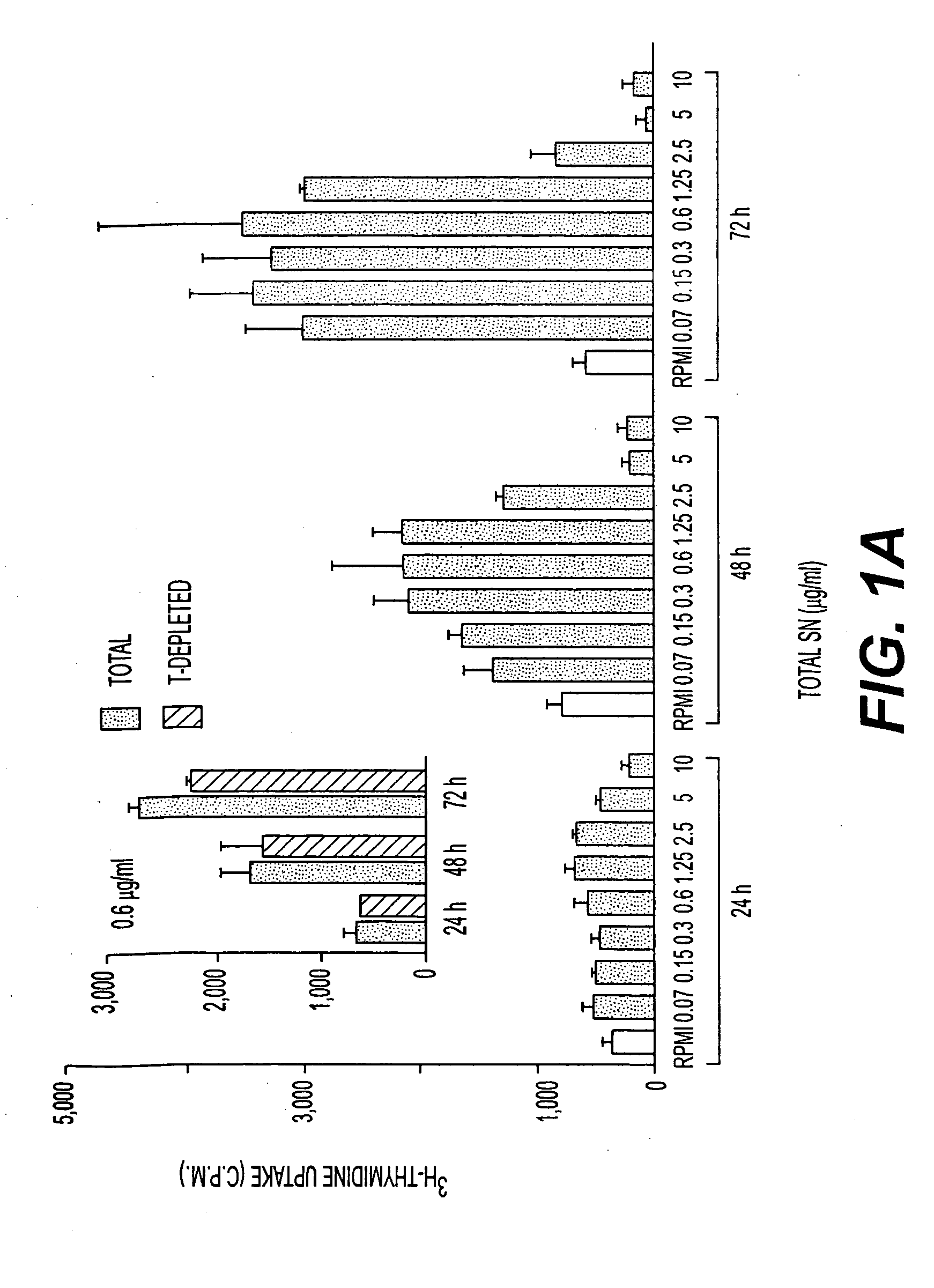
[0218] 40 liters of culture supernatants from metacyclic forms, maintained for an additional 96 h at 37° C., were concentrated by vacuum dialysis and dialyzed against buffer A. HPLC was performed using a weak anion exchanger column POROS HQ-10 (Perspective Biosystems) at a flow rate of 1 ml / min according to the following program: a) 10 min with buffer A; b) 30 min linear gradient from buffer A to B; c) 5 min linear gradient from buffer B to C; and d) 5 min with buffer C. One ml fractions were collected, frozen at −80° C., lyophilized and reconstituted in H2O or in non-supplemented RPMI medium for in vitro proliferation assays. (Buffers used: A: 5 mM NH4-acetate, pH 8. B: 1M NH4-acetate, pH 8. C: 1M NaCl / 1M NH4-acetate, pH 8). Fractions 1 ml in volume were collected, frozen at −80° C., lyophilized and reconstituted in water or in non-supplemented RPMI medium for in vitro proliferation assays. SDS-PAGE analysis used standard techniques.
example 3
Generation of Peptides and Amino Acid Sequence Analysis
[0219] HPLC fractions 22 and 23 were pooled and fractionated by 8% SDS-PAGE. After amino black staining, the 45 kDa protein band was cut out, in-gel digested with trypsin, and submitted to reverse phase HPLC to separate peptides. Automated Edman degradation sequence analysis was performed in the Laboratoire de Microséquençage de Protéines of the Pasteur Institut.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com