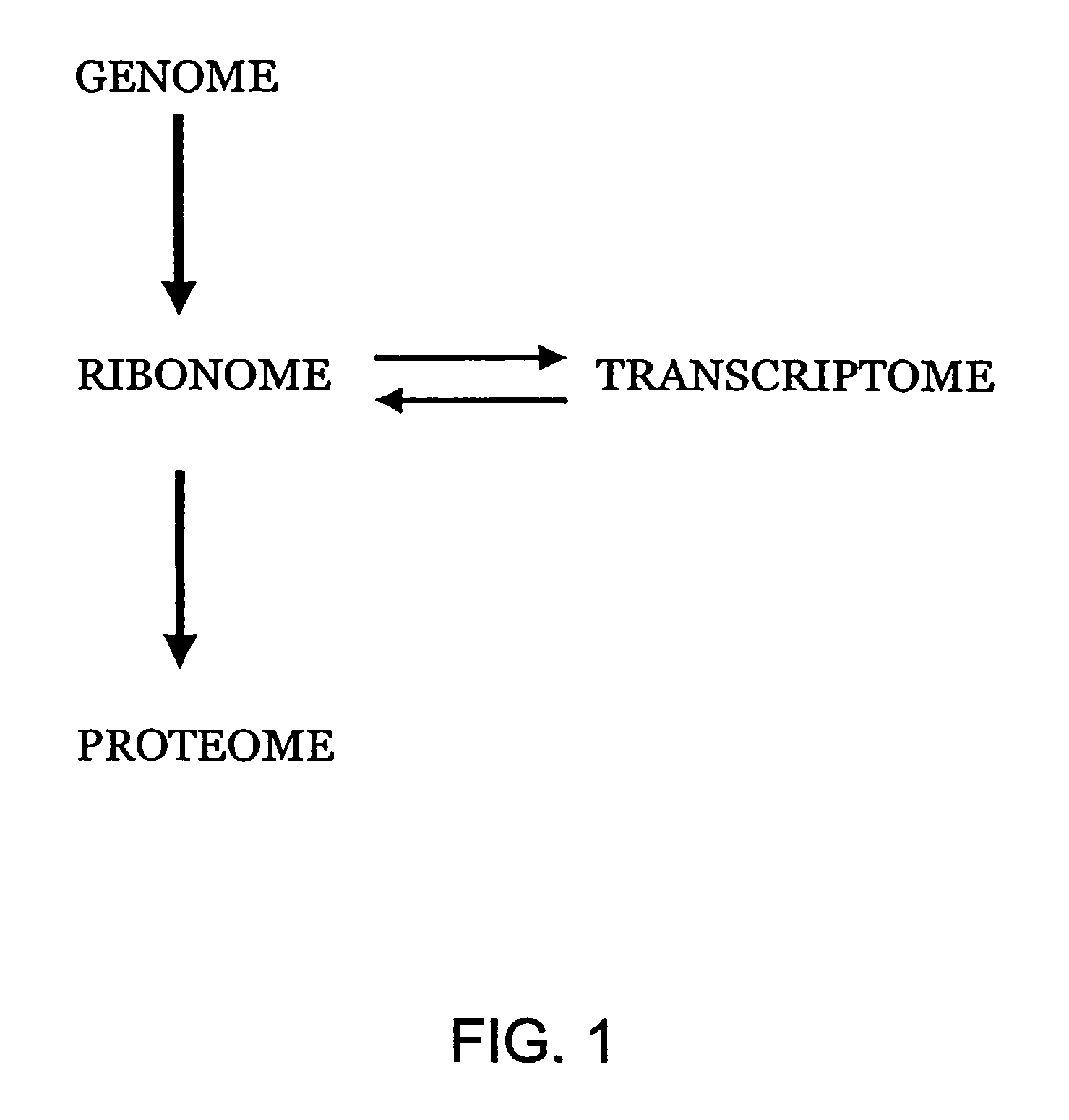
Methods for identifying functionally related genes and drug targets
a functionally related gene and drug target technology, applied in the field of methods for identifying functionally related genes and drug targets, can solve problems such as the inability of the vitro method to refl
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
RNase Protection in a Multiprobe System
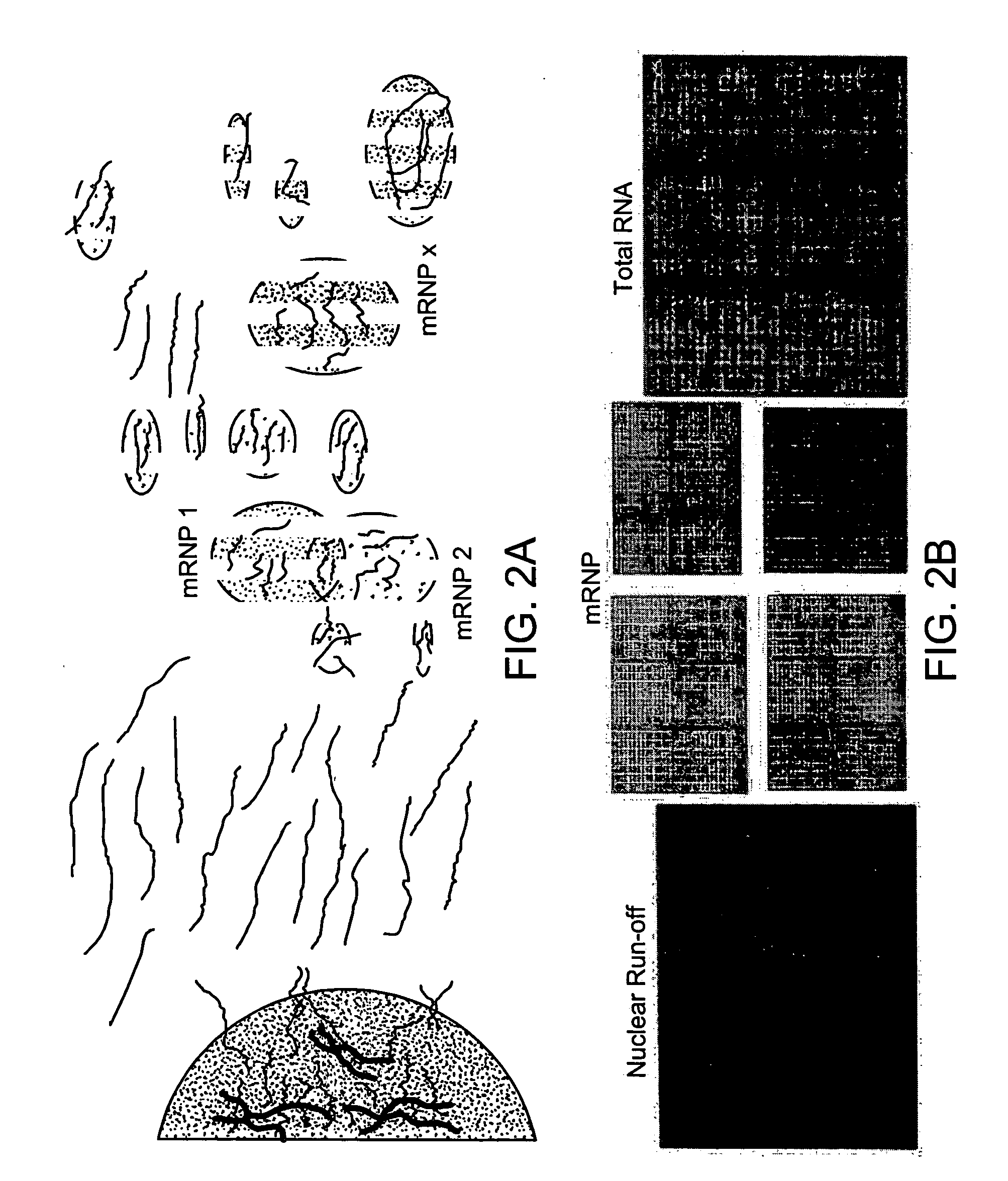
[0093] A multiprobe RNase protection assay was used to rapidly optimize the immunoprecipitation of several endogenous mRNP complexes containing different RNA binding proteins. In the multiprobe system, many mRNAs associated with an mRNP complex can be assayed in a single lane of a polyacrylamide gel.
[0094] Cell Culture and Transformation. Murine P19 embryonal carcinoma cells were obtained from the American Type Culture Collection (ATCC, Manassas, Va.) and maintained in monolayer culture using α-Minimum Essential Medium Eagle (MEM) without phenol red (Gibco BRL 41061-0291) (Invitrogen, Carlsbad, Calif.) supplemented with 7.5% Bovine Calf Serum (BCS), 2.5% Fetal Bovine Serum (FBS) (Hyclone, Logan, Vt.) and 100 U of Penicillin / Streptomycin. Cells were grown in tissue culture flasks or plates that had been pre-coated with 0.1% gelatin (Sigma Chemicals, St. Louis, Mo.) and removed prior to use. Monolayer cell cultures were maintained in 5% CO2 at ...
example 2
Identification of mRNA Subsets Associated with RNA Binding Proteins En Masse Using cDNA Arrays
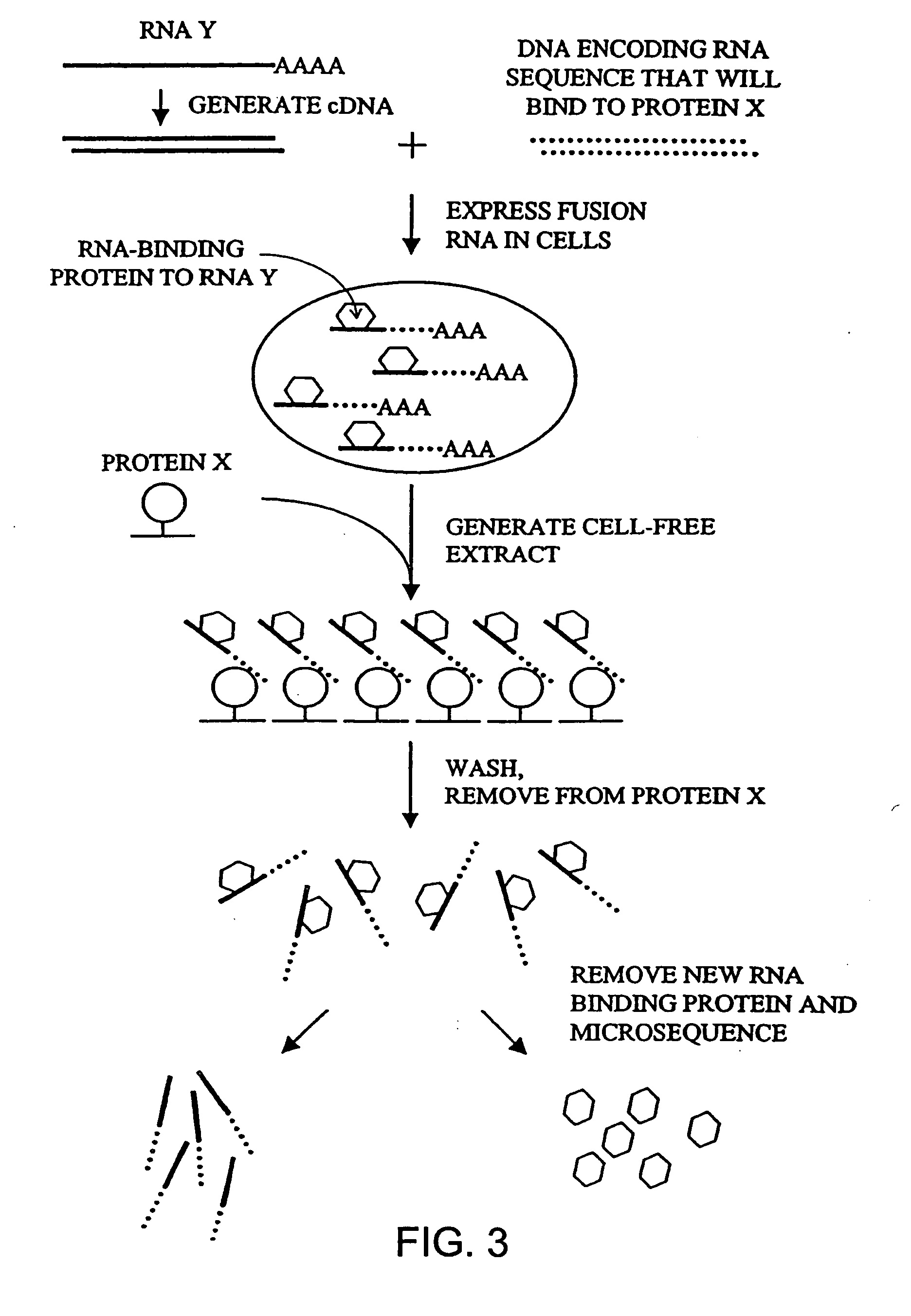
[0101] A cDNA array (FIG. 5) was used to detect an mRNA subset without amplification or iterative selection.
[0102] Antibodies. Monoclonal anti-gene 10 (g10) antibodies and polyclonal sera reactive with the proteins were produced as previously described. Antibody against 5′ cap binding protein (elF-4E) was obtained from Transduction Laboratories (San Diego, Calif.). Antibodies reactive with Poly A-binding protein (PABP) were obtained from McGill University (Canada).
[0103] Cell Culture and Differentiation. Transgenic cells were prepared as described in Example 1. Cells were treated with retinoic acid (RA) to induce neuronal differentiation as follows: 5×105 p19 cells were placed on a 60 mm petri dish (Fisher Scientific, Pittsburgh, Pa., Number 8-757-13A) with 0.5 μM RA (Sigma Chemicals, St. Louis, Mo., Number R2625). After two days, 25% of the cells that had formed into clumps were removed...
example 3
Alterations in mRNP Complexes in Response to Retinoic Acid
[0110] Because HuB is predominantly a neuronal protein believed to play a role in regulating neuronal differentiation, studies were conducted to investigate whether the mRNA population found in HuB mRNP complexes changes in response to RA, a chemical inducer of neuronal differentiation. HuB-transfected P19 cells were treated with RA to induce the onset of neuronal differentiation, HuB mRNP complexes were immunoprecipitated, and then associated mRNAs were identified on cDNA arrays as described in Examples 1 and 2. Comparison of the mRNA profiles extracted from the HuB mRNP complexes before and after RA treatment revealed that eighteen mRNAs were either exclusively present or greatly enriched (four-fold or greater) in RA-treated HuB mRNPs (FIGS. 7A and 7B). In addition, three mRNAs (T-lymphocyte activated protein, DNA-binding protein SATB1, and HSP84) decreased in abundance by four-fold or greater in response to RA treatment (...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com