Gene expression analysis using array with immobilized tags of more than 25 bp (SuperSAGE-Array)
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
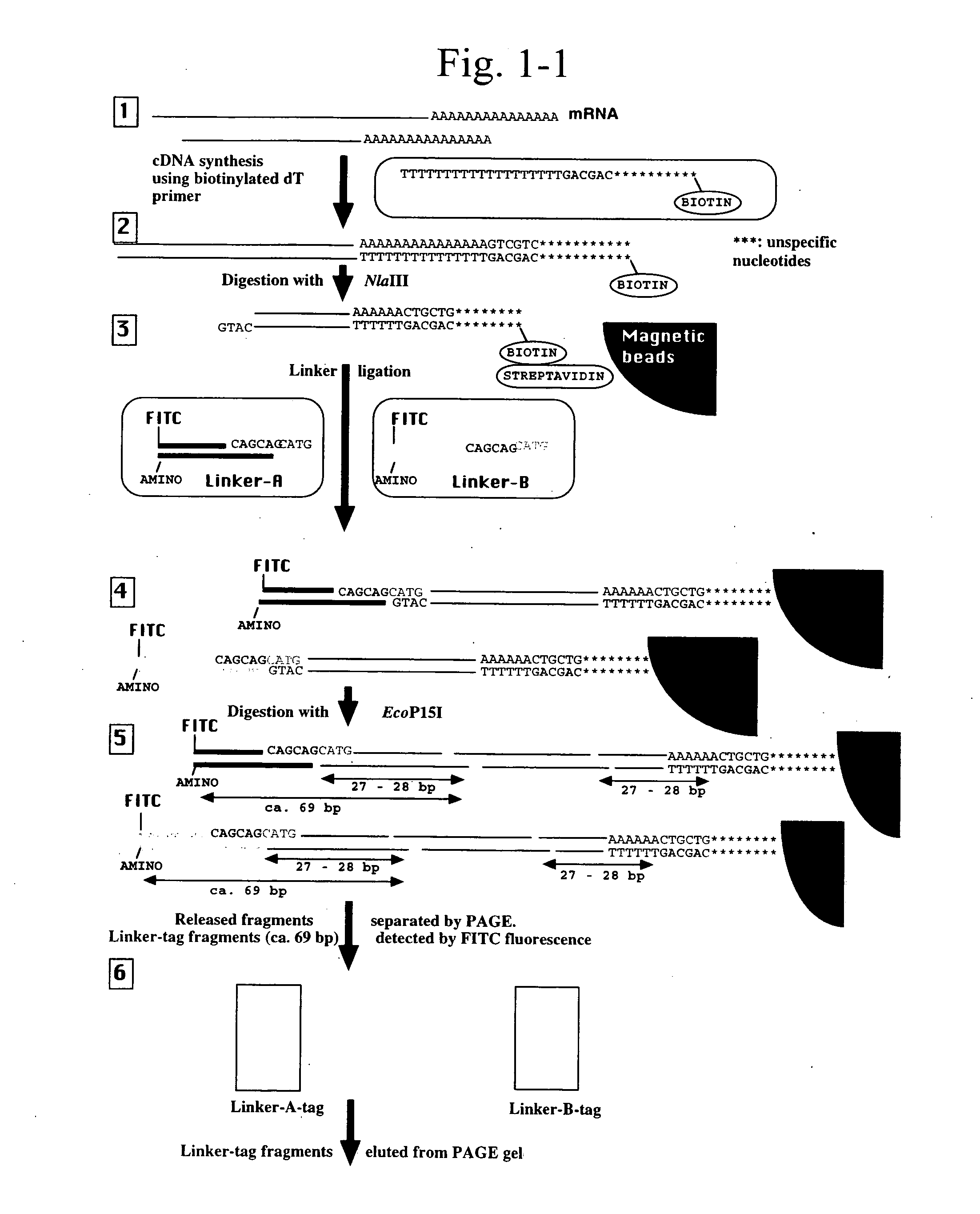
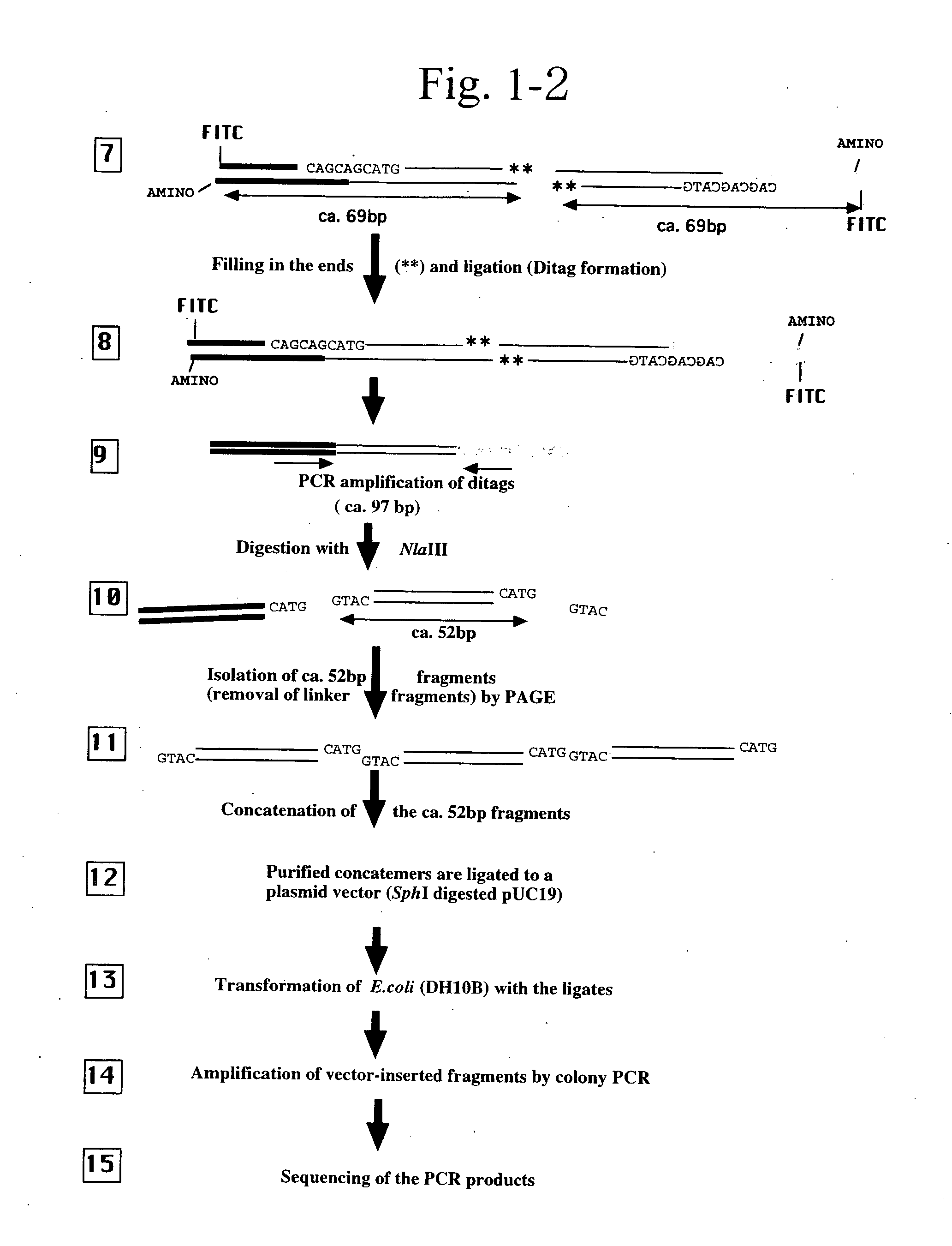
1. Material and Method
1) Preparation of RNA
[0055] For rice SuperSAGE and oligoarray systems, rice leaves (variety: Yashiromochi) and suspension-cultured cells (variety: Kakehashi) were prepared. For the oligoarray system, mRNA was extracted from the rice (variety: Yashiromochi) and the cultured cells (variety: Kakehashi) 1 month after sowing using an mRNA Purification Kit (Amersham Pharmacia).
[0056] For Nicotiana benthamiana SuperSAGE and oligoarray systems, leaves into which Agrobacterium containing the following plasmids had been injected were prepared. Two days after the Agrobacterium injection, Nicotiana benthamiana leaves were treated with dexamethasone (DEX), and mRNA was extracted using the mRNA Purification Kit (Amersham Pharmacia) 4 hours later.
2) Plasmids
[0057] NbCD1 (JP Patent Publication (Unexamined) No. 2005-278634) and NbCD3 cDNA (JP Patent Publication (Unexamined) No. 2005-245251), which had been isolated by the present inventors in the past, were used. A bina...
example 2
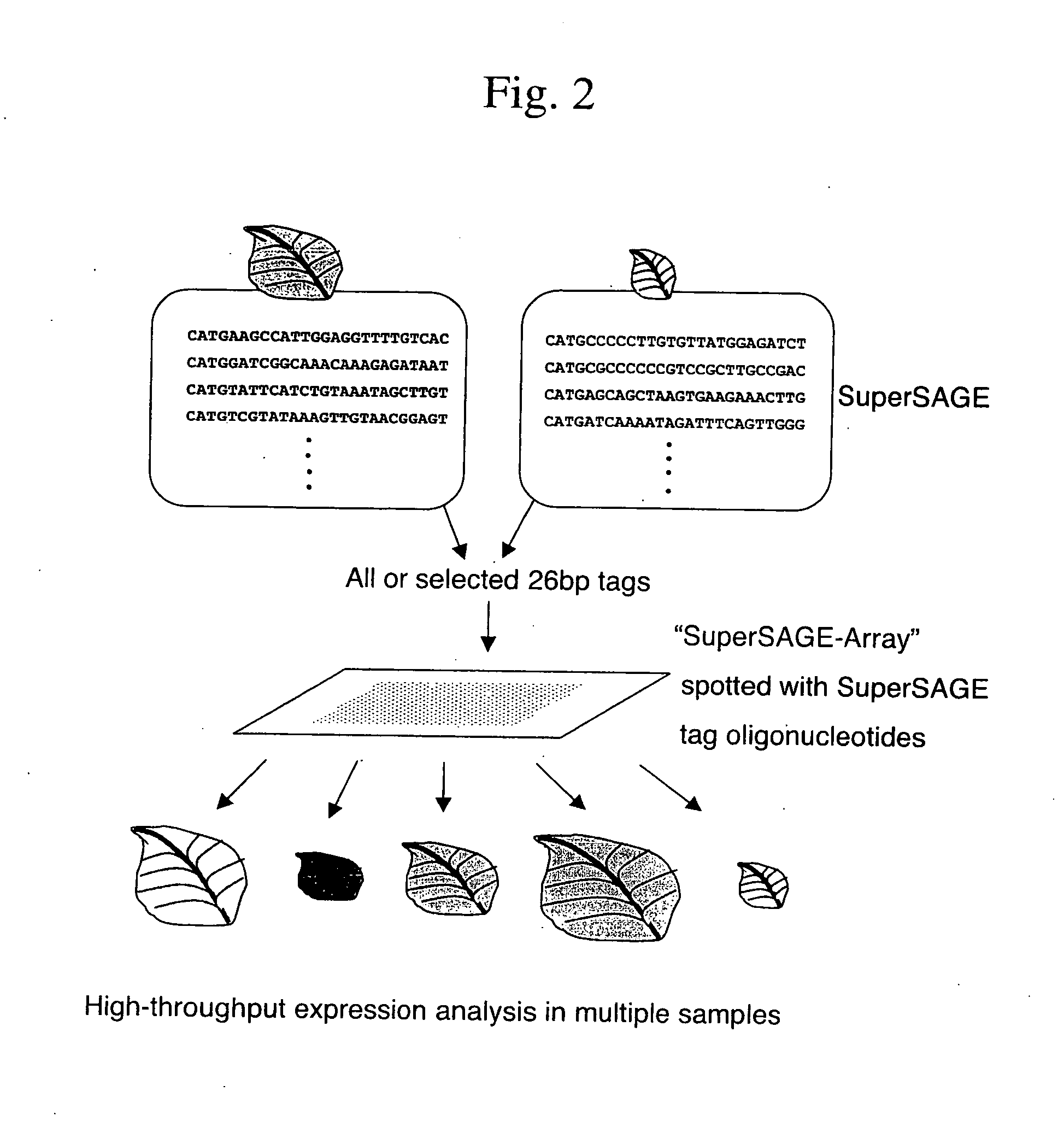
[0093] In Example 1, model rice plants were subjected to expression analysis using an array with 41 SuperSAGE tags immobilized thereon, and non-model Nicotiana benthamiana plants were subjected to expression analysis using an array with 154 SuperSAGE tags immobilized thereon. In both cases, the results of analysis were very consistent with the results of expression analysis via SuperSAGE. In this example, an array with 1,000 SuperSAGE tags immobilized thereon was prepared for rice, and the results of expression analysis via SuperSAGE-Array were compared with those via SuperSAGE.
[0094] In accordance with the procedure of Example 1, mRNAs were extracted from rice leaves (variety: Yashiromochi) and cultured cells of rice (variety: Kakehashi) to prepare SuperSAGE libraries. From these libraries, 1,000 SuperSAGE tag sequences were selected. Among them, 78 tags represented equally expressed genes, 438 tags were more prevalent in leaves and 484 tags were more abundant in suspension-cultur...
example 3
[0098] Among the tags that were found to be expressed at high levels in all of the NbCD1- and NbCD3-overexpressing Nicotiana benthamiana leaves by the SuperSAGE-Array-based expression analysis in Example 1, 5 tags showing no sequence matches to known cDNA or EST were selected (NbCD3U14, 20, 25, 32, and 40), and identification of the genes corresponding thereto was attempted.
[0099] Full-length sequences of the tags were determined by the 3′-RACE and 5′-RACE methods in the following manner. As a template, RNA was isolated from NbCD3-overexpressing Nicotiana benthamiana leaves, and the RNA was flanked by adaptor sequences to synthesize cDNA. Based on the SuperSAGE tag sequences, a gene specific PCR primer and a primer complementary to the adaptor sequence were used to amplify a partial cDNA fragment from template cDNA. A primer was prepared based on the 5′-sequence of the resulting fragment, and the resulting primer and the adaptor primer (i.e., a primer complementary to the adaptor s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com