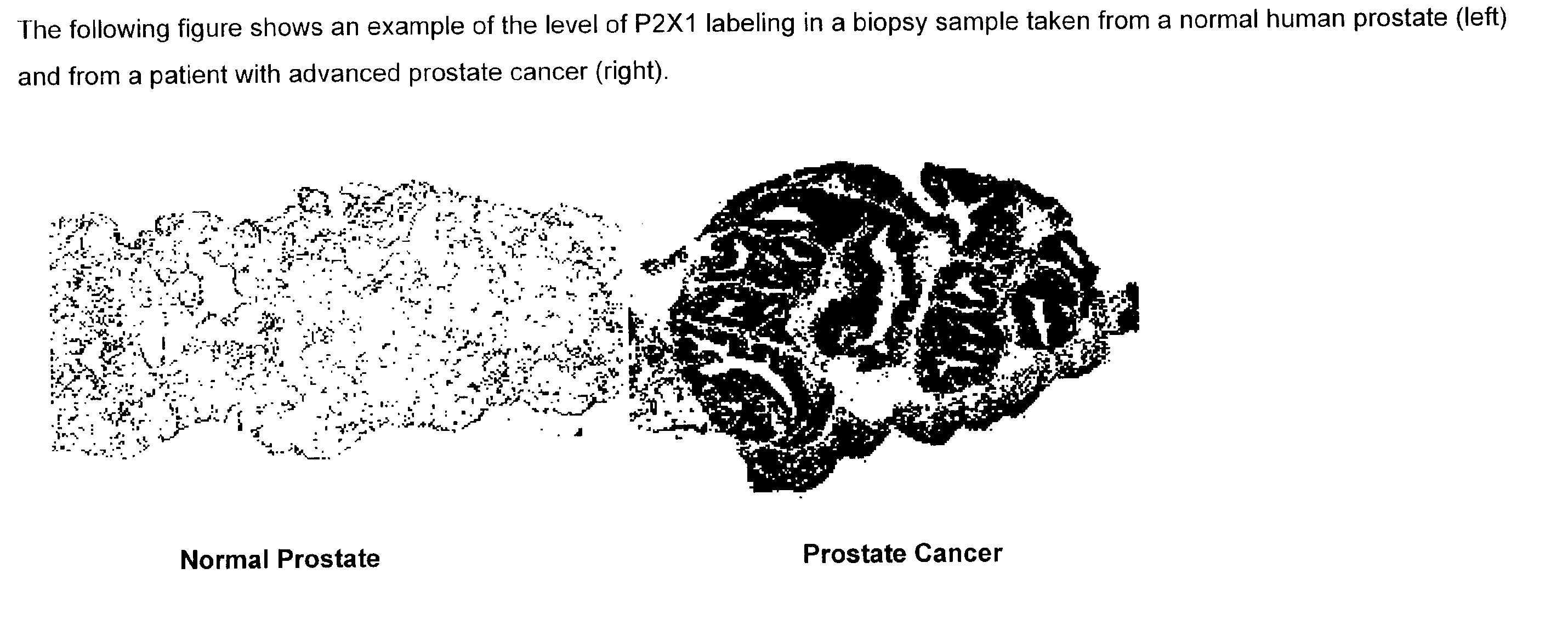
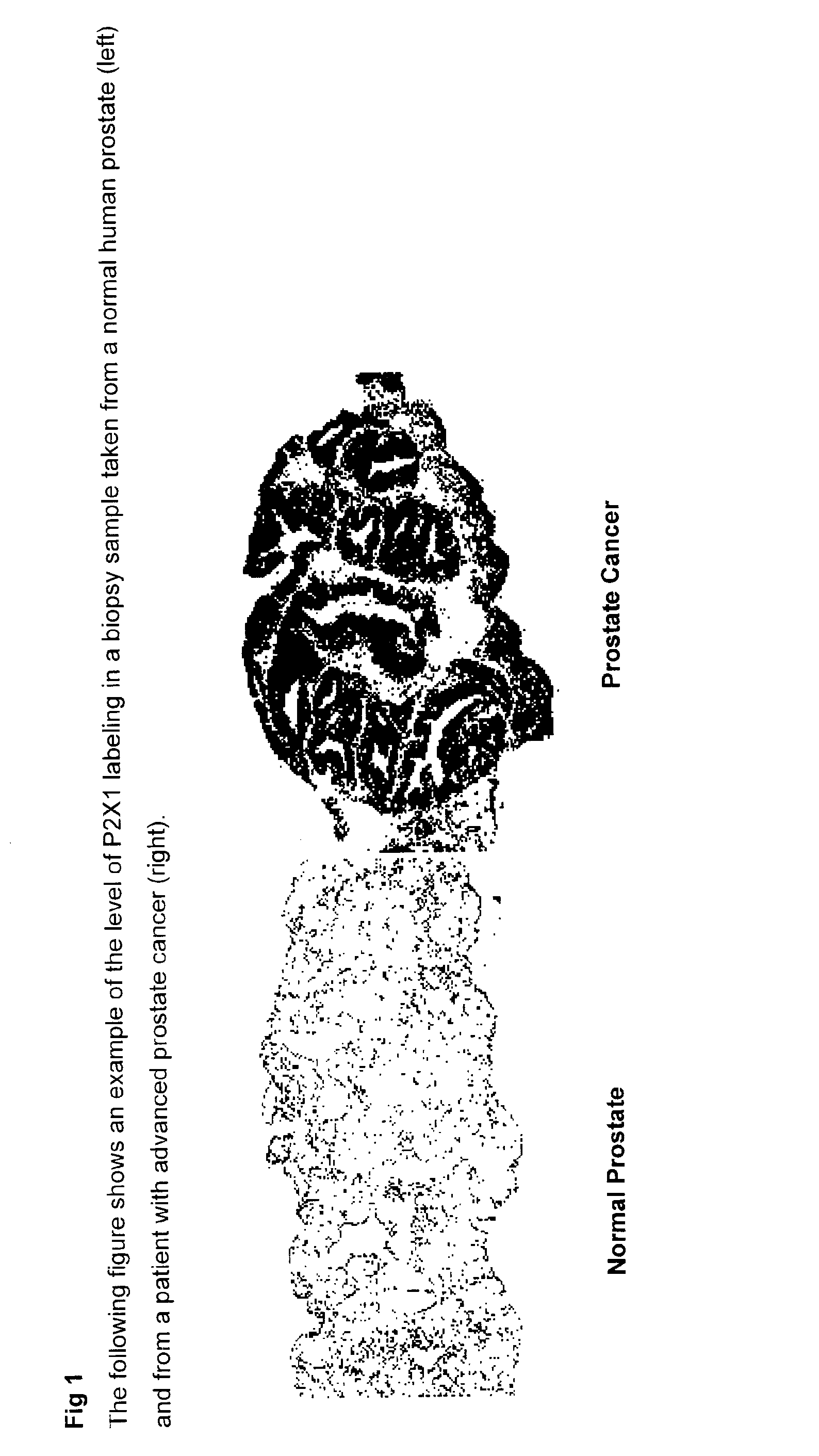
Method for identifying pre-neoplastic and/or neoplastic states in mammals
a preneoplastic and/or neoplastic state technology, applied in the field of mammals preneoplastic and/or neoplastic states identification, can solve the problems of few diagnostic methods, inconvenient diagnosis, and inability to accurately predict the age of the patient,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Immunohistochemical Procedure
[0042] The immunohistochemical method used in this study was adapted from Barclay [31]. Sections with a thickness of 8 μm were cut from unfixed, frozen tissue using a Reichert Jung 2800 Frigocut cryotome. Sections were air dried at room temperature for 1 hour, fixed for 12 hours in acetone at −20° C. and air dried at room temperature for 1 hour prior to antibody labelling. They were then incubated at room temperature with one of either rabbit or sheep anti-P2X1, P2X2, P2X3, P2X4, P2X5, P2X6 or P2X7 antibody. After washing, sections were then incubated in the secondary antibody; a 1:30 dilution of HRP-conjugated goat anti-rabbit secondary antibody (Dako) for 30 mins for rabbit primaries and HRP-conjugated goat anti-sheep secondary antibody (Dako) for sheep primaries. Slides were again rinsed and then immersed in 15% diaminobenzidine tetrahydrochloride (DAB—Sigma) for 10 minutes. Sections were rinsed, air dried and mounted in DPX (Merck). Control slides w...
example 2
Antibody Production
[0043] The consensus sequences of the rat P2X1 [32], P2X2 [33], P2X3 [34], rat P2X4 [35], rat P2X5 [36], rat P2X6 [36], rat P2X7 [37], human P2X7 [38], human P2X7 [39], human P2X3, [40], human P2X4 [41] and human P2X5 [42] cloned receptors were examined for suitable epitopes following the approach adopted in Hansen et al. [15]. The non-homologous epitopes corresponding to the segment Lys 199-Cys217 used in rat P2X1 were utilised in rat P2X3, rat P2X6 and rat P2X7. Variations were applied to rat P2X4 which used the sequence Ile235-Gly251 to which was attached a C-terminal Cys residue for cross-linking to a 6 kDa diphtheria toxin domain. The P2X2 epitope was selected from a region within the C1 domain [15], Cys130-Gly153. The rat P2X5 epitope was selected from a region closer to the second transmembrane domain but still extracellular (Lys314-Ile333 to which was added a C-terminal Cys also for conjugation). Although largely homologous with rat P2X4, cross-labelling ...
example 3
Specificity of Antibodies
[0045] Each of the P2X antisera used has been shown to possess similar distributions in many cases but with distinctly different distributions in other cases indicating that the antisera do not lack specificity. Specificity was demonstrated by affinity purification of the sera against the cognate peptides. To further verify antibody specificity, individual antibody such as the antibody to P2X1 was added to cells transfected with the corresponding P2X1 cDNA in the presence and absence of a 10 mM concentration of the P2X epitope. Immunolabelling and confocal imaging of the tranfected Xenopus oocytes demonstrated that the expressed P2X1 is located, as expected, within the cell membrane and the presence of a 10 mM concentration of the cognate peptide as an absorption control resulted in the blocking of P2X1 staining [18].
[0046] Individual specificity of all other antibodies has been similarly demonstrated.
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| volumes | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com