sCD Fingerprints
a fingerprint and histopathological technology, applied in the field of scd fingerprints, can solve the problems of insufficient accuracy of histopathological approach, high cost, impracticality, etc., and achieve the effect of complicating the cd fingerprint obtained
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
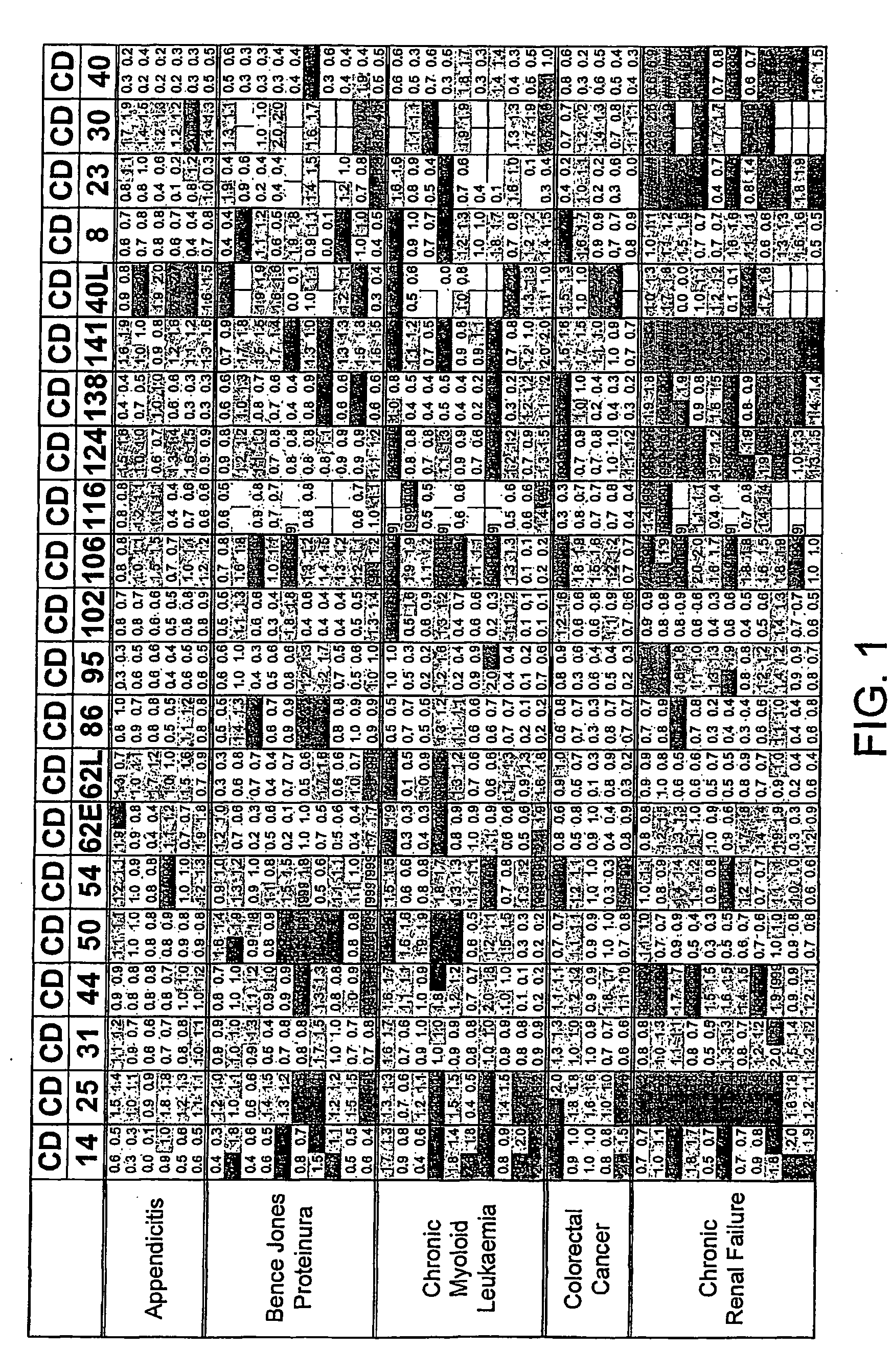
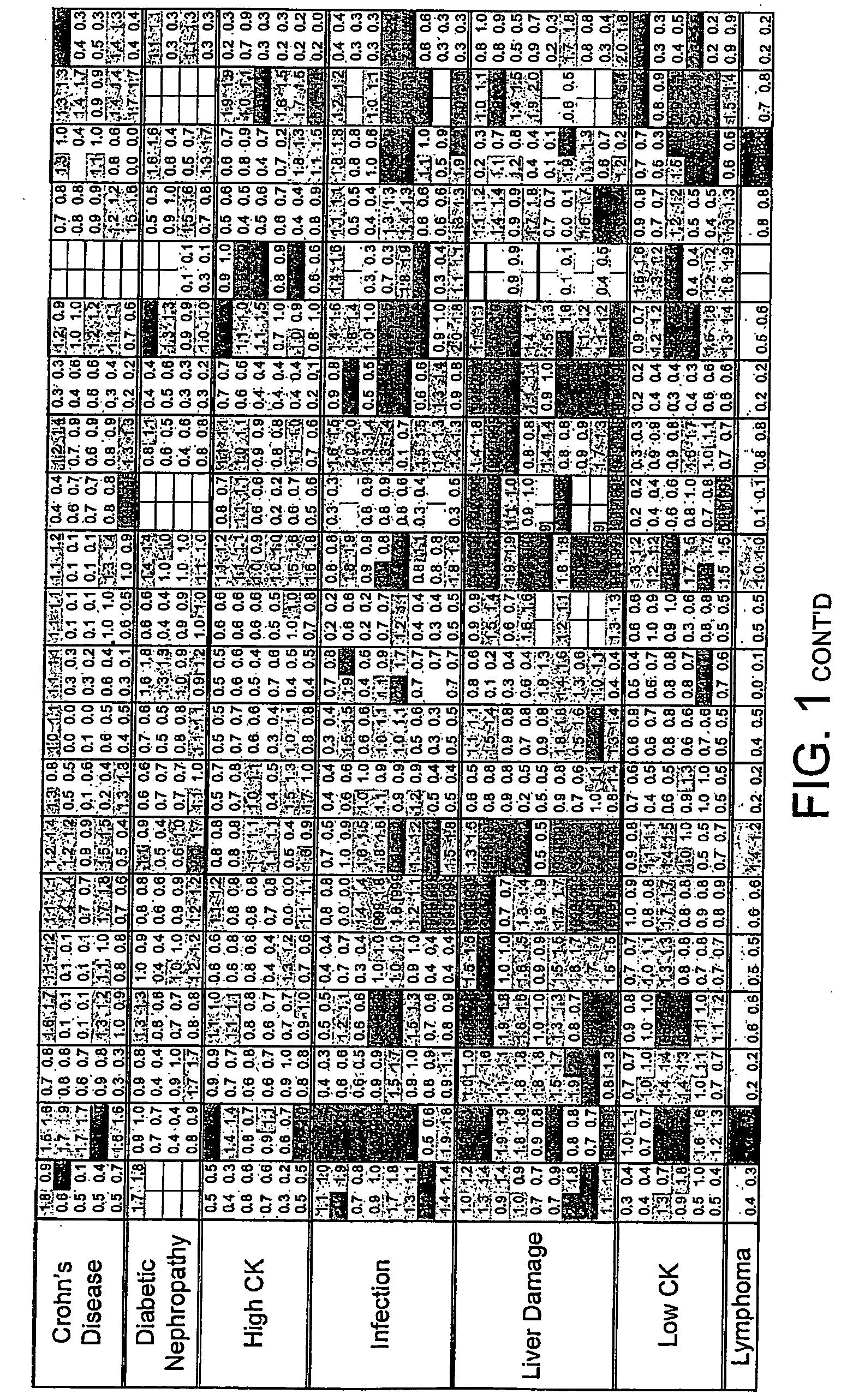
FIG. 1. Disease Groups. Multiples of ULN All sCD's included
[0215]Two values obtained (CD40L and CD30) for the individual, classified as normal, with a suspected drug overdose were omitted from the calculation of the upper limit of normal. The dilution factor for each sCD was fixed throughout the study. The results obtained are those of the diluted sample and have not been multiplied by the dilution factor. The absolute value of each data point was divided by the upper limit of normal (ULN) as defined above. Where the absolute value was greater than the dynamic range of the assay the result [9999] was recorded.
[0216]The limits indicated by each point are:[0217]Green ≦1×ULN[0218]Blue 1-2×ULN[0219]Red >2×ULN[0220]A white block indicates no data available.
example 2
FIG. 2 Remove all sCD's that Appear not to Discriminate from the Normals (sCD's 21; 102; 117; 126; 130; 26; 44v5; 44v6; 62P)
[0221]To simplify the diagram the above sCD plots were removed.
[0222]The data suggests (FIG. 1.) that the concentration of some of these sCD may actually be lowered in disease. As we initially worked on the premise that there would be over-expression of these molecules in disease, samples have been diluted optimally to focus on high, rather than low concentrations.
example 3
FIG. 3. Disease Groups. Mode of Response for Remaining 20 sCD's
[0223]To simplify the data further the modal response for each disease group has been plotted. As the lymphoma and “oligoclonal-banding positive” group contain only a single subject, they have been omitted. Where there is no clear mode, both responses have been shown.
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com