Methods for producing a protein using an avian lysozyme promoter
a technology of lysozyme and promoter, which is applied in the direction of viruses/bacteriophages, immunoglobulins, peptides, etc., can solve the problems of limited success and attendant costs of maintaining individual animals, and achieve the effect of reducing the positional variation of transgenic avians
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of Lysozyme Promoter Plasmids
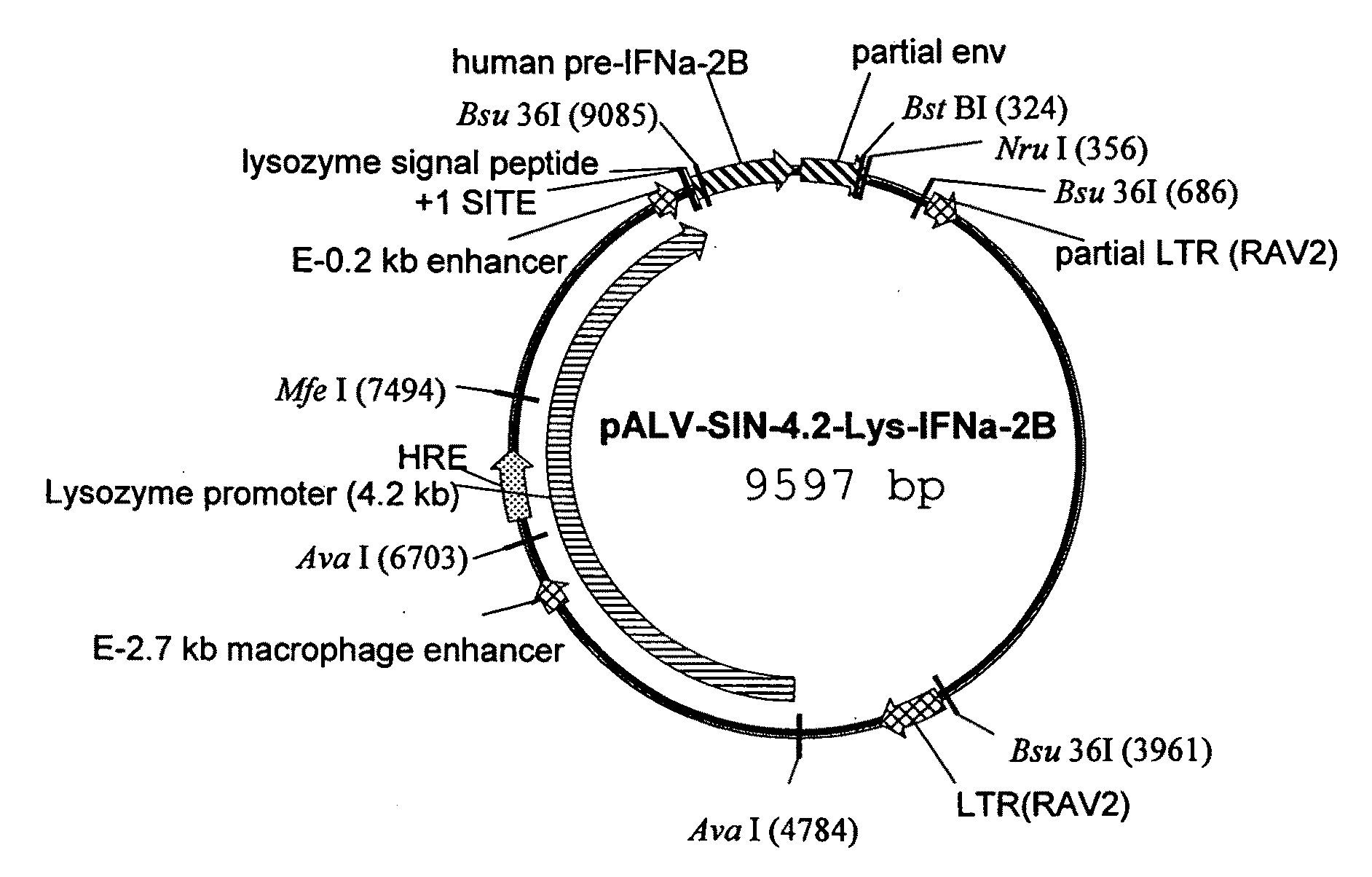
[0191]The chicken lysozyme gene expression control region was isolated by PCR amplification. Ligation and reamplification of the fragments thereby obtained yielded a contiguous nucleic acid construct comprising the chicken lysozyme gene expression control region operably linked to a nucleic acid sequence optimized for codon usage in the chicken (SEQ ID NO: 66) and encoding a human interferon α2b polypeptide optimized for expression in an avian cell.
[0192]White Leghorn Chicken (Gallus gallus) genomic DNA was PCR amplified using the primers 5pLMAR2 (SEQ ID NO: 1) (see FIG. 1) and LE-6.1 kbrev1 (SEQ ID NO: 2) in a first reaction, and Lys-6.1 (SEQ ID NO: 3) and LysE1rev (SEQ ID NO: 4) as primers in a second reaction. PCR cycling steps were: denaturation at 94° C. for 1 minute; annealing at 60° C. for 1 minute; extension at 72° C. for 6 minutes, for 30 cycles using TAQ PLUS PRECISION™ DNA polymerase (Stratagene, LaJolla, Calif.). The PCR products...
example 2
Construction of Plasmids which Contain the 3′ Lysozyme Domain
[0195]The plasmid pAVIJCR-A 115.93.1.2 was restriction digested with FseI and blunt-ended with T4 DNA polymerase. The linearized, blunt-ended pAVIJCR-A 115.93.1.2 plasmid was then digested with XhoI restriction enzyme, followed by treatment with alkaline phosphatase. The resulting 15.4 kb DNA band containing the lysozyme 5′ matrix attachment region (MAR) and −12.0 kb lysozyme promoter driving expression of a human interferon was gel purified by electroelution.
[0196]The plasmid pIIIilys was restriction digested with MluI, then blunt-ended with the Klenow fragment of DNA polymerase. The linearized, blunt-ended pIIIilys plasmid was digested with XhoI restriction enzyme and the resulting 6 kb band containing the 3′ lysozyme domain from exon 3 to the 3′ end of the 3′ MAR was gel purified by electroelution. The 15.4 kb band from pAVIJCR-A115.93.1.2 and the 6 kb band from pIIIilys were ligated with T4 DNA ligase and transformed i...
example 3
Sequencing Reactions
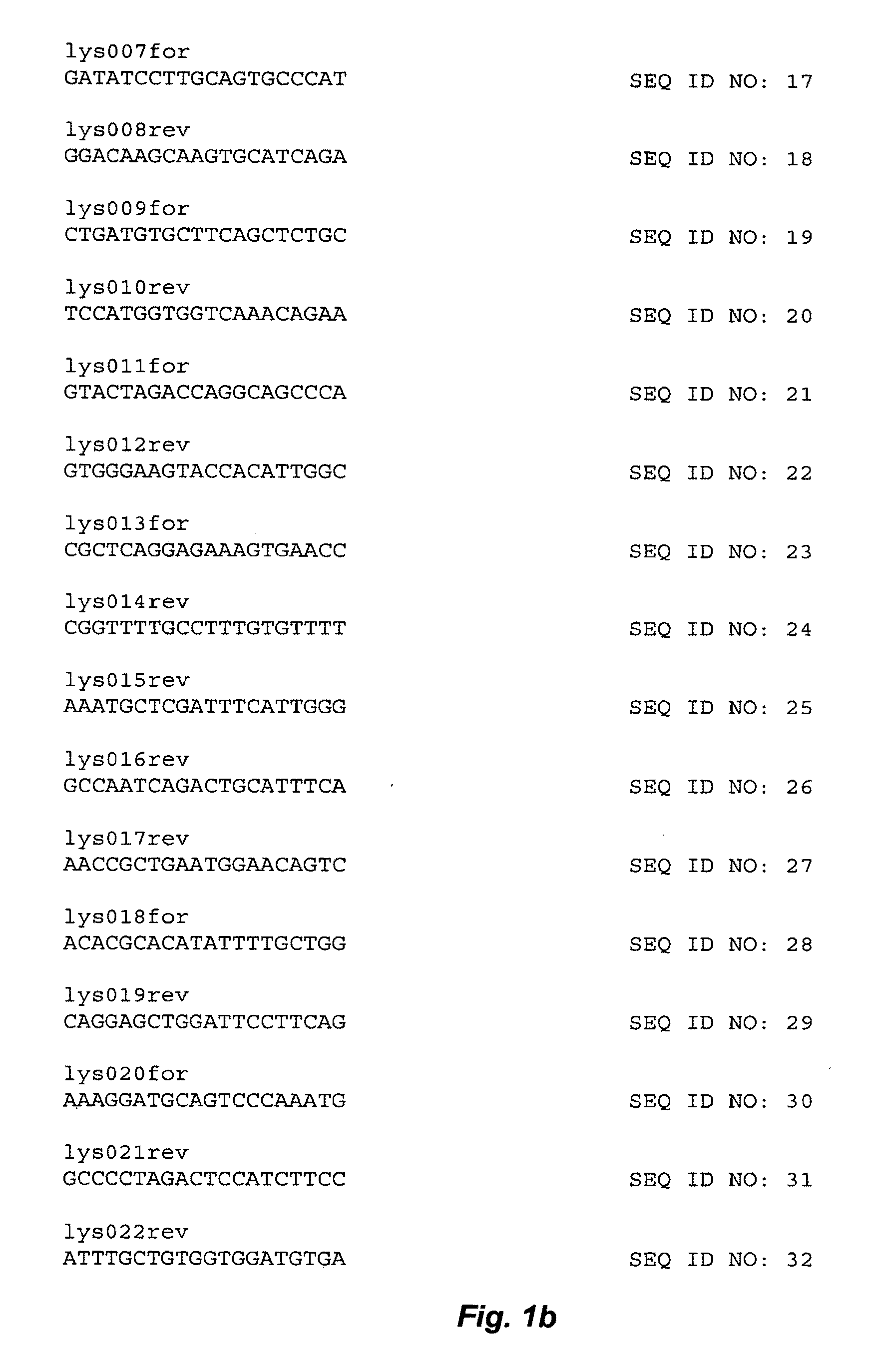
[0197]Plasmid DNA (pAVIJCR-A115.93.1.2) produced as described in Example 1 was purified with QIAGEN™ columns (Qiagen, Valencia, Calif.). Sequencing reactions were performed according to the Applied Biosystems (Foster City, Calif.) protocol for BIGDYE™ Terminators, version 2.0, using an ABI 373 Stretch sequencer. Sequencing primers used are listed in FIG. 1, and a schematic diagram illustrating the sequencing reactions using the different primers is shown in FIG. 2. Sequence data was analyzed with SEQUENCHER™ software, version 4.0 (Gene Codes Corp., Ann Arbor, Mich.).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com