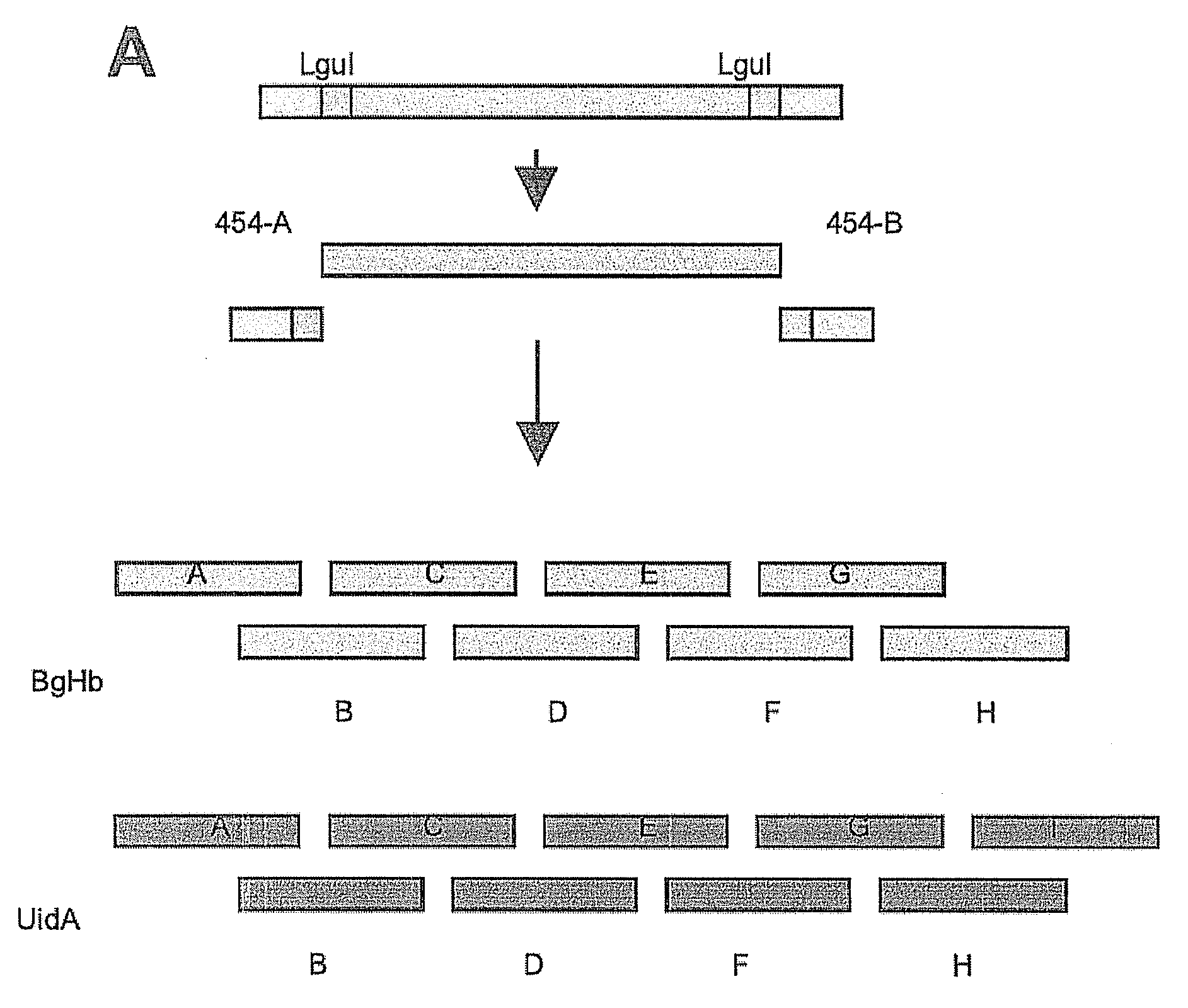
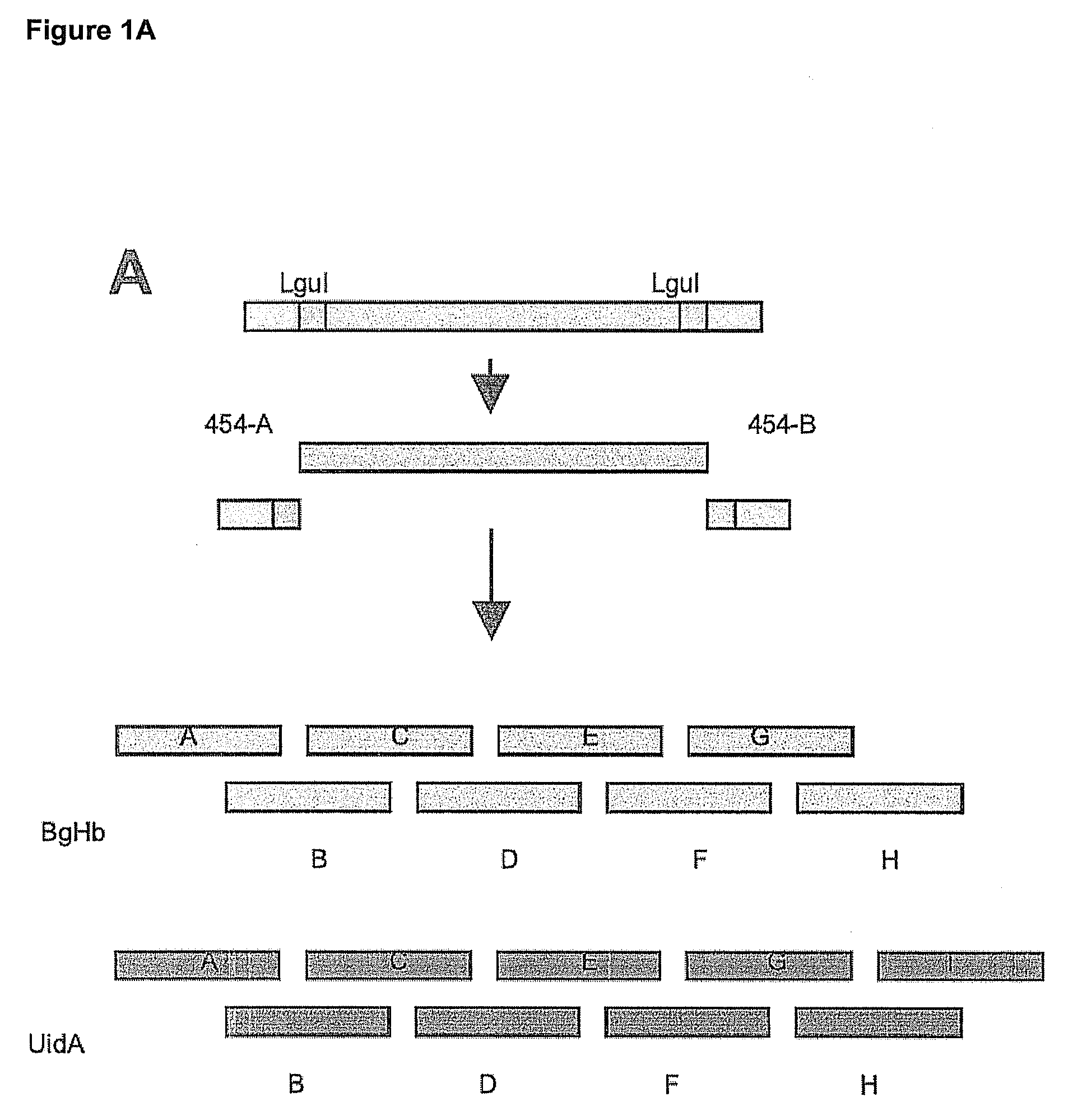
Synthesis of sequence-verified nucleic acids
a nucleic acid and sequence verification technology, applied in the field of synthetic nucleic acids, can solve the problems of not finding such widespread application, and achieve the effects of high error rate and cost, high quality, and high impact of oligonucleotide input error ra
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
7.1 Example 1
Preparative Next Generation Sequencing
High Parallel In Vitro Cloning and Sequencing for Gene Synthesis and Library Preparation
7.1.1 Summary
[0412]The major cost factors of synthetic gene production are oligonucleotides as starting material for gene assembly, and the subsequent screening and selection of correct parts from a mixture of correct and defective gene fragments resulting from errors in oligonucleotide synthesis(1). While microarrays and especially photo-programmable microfluidic chips(2,3) represent an effective tool to decrease the cost for oligonucleotide production by orders of magnitude), the overall production cost for synthetic genes remains high due to the need for cloning and sequencing post-synthesis. Here we describe a proof of concept for a high throughput retrieval of clonal DNA with known sequence from a next generation sequencing (NGS) platform(5). This technology will reduce efforts for quality control, screening and selection of DNA significantl...
example 2
7.2 Example 2
[0439]Analysis of sequence data of mega cloned DNA fragments.
[0440]319 clonal beads from a picotiter plate (PTP) of a 454 sequence instrument were picked. Each bead carried a different sequence from a PTP. The bead-associated nucleic acids were amplified and pooled. The pooled nucleic acid sequences were sequenced on an Illumina GAII instrument to analyse the sequence verification.
[0441]The results are shown in FIG. 10.
[0442]For 304 beads the correct sequence was found with at least 20× coverage which should be regarded as solid proof that the correct sequence has indeed been picked. With a sequence length of 40 by fragment, the overall DNA length from this run was at least 1260 by error-free DNA.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| constant temperature | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com