Cultures with Improved Phage Resistance
a technology of phages and cultures, applied in the field of cultures with improved phage resistance, can solve the problems of occupying a large space and equipment, phage contamination, and phage contamination, and achieve the effect of reducing the degree of homology to the spacer, reducing or decreasing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Manipulation of Phage-Specific Spacers
[0481]In this Example, experiments conducted to manipulate phage-specific spacers are described. In some experiments, a phage specific spacer was inserted into an existing, functional CRISPR to provide resistance to the corresponding phage are described. The bacterial strain used was Streptococcus thermophilus ST0089 and the phage was phage 2972. S. thermophilus ST0089 is an industrially important strain used in the manufacture of yogurt. It is genetically amenable to manipulation, and susceptible to the well-known virulent phage 2972.
[0482]The CRISPR loci were determined in strain ST0089. This was determined preferentially by sequencing the entire genome of ST0089. Alternatively, the CRISPR loci are identified via PCR using primer sets with sequences identical to S. thermophilus CRISPR elements previously identified.
[0483]Once identified, the CRISPR loci sequences were determined as well as the proximal regions containing the relevant cas genes...
example 2
Integration of CRISPR Spacer(s)
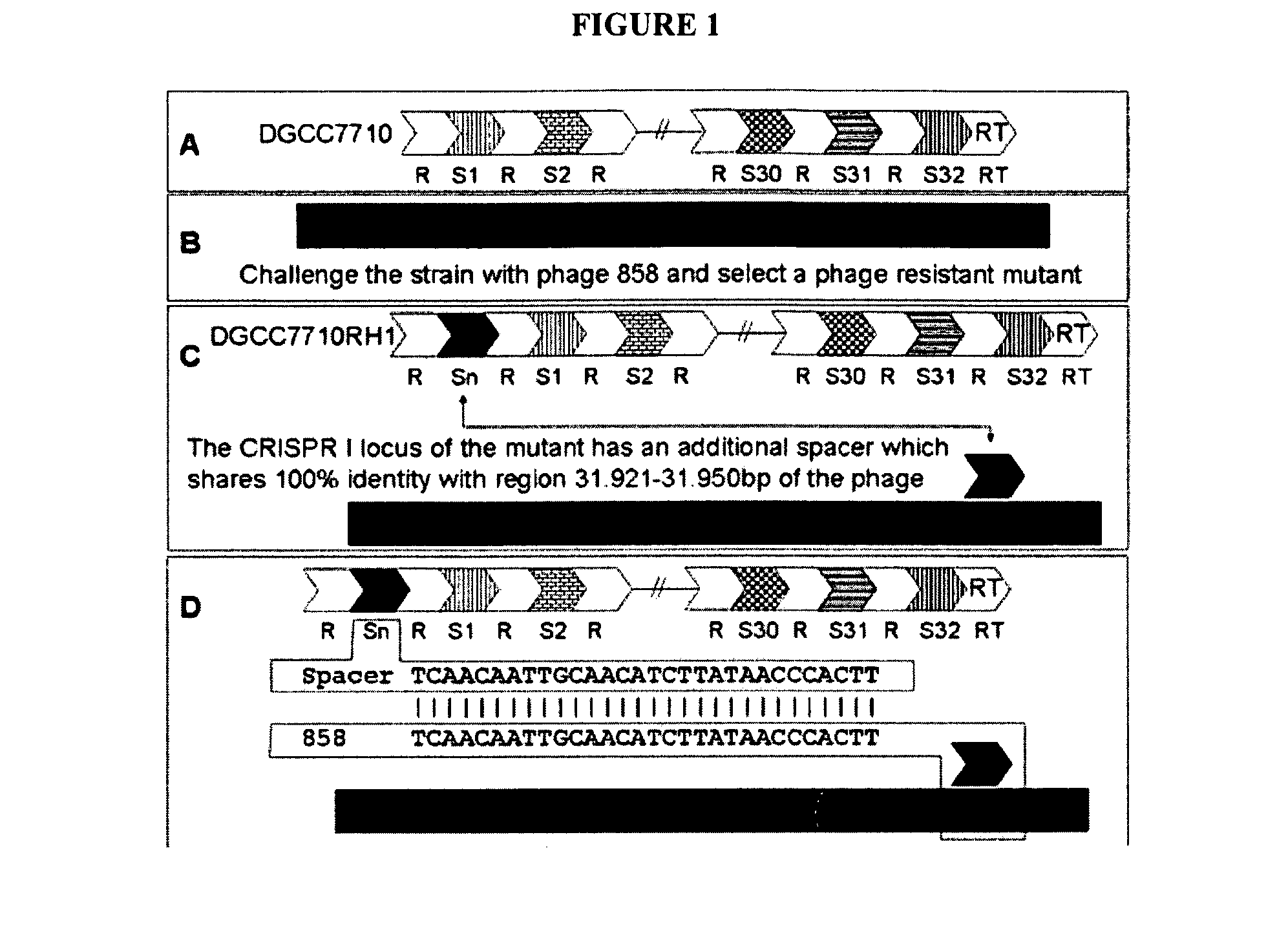
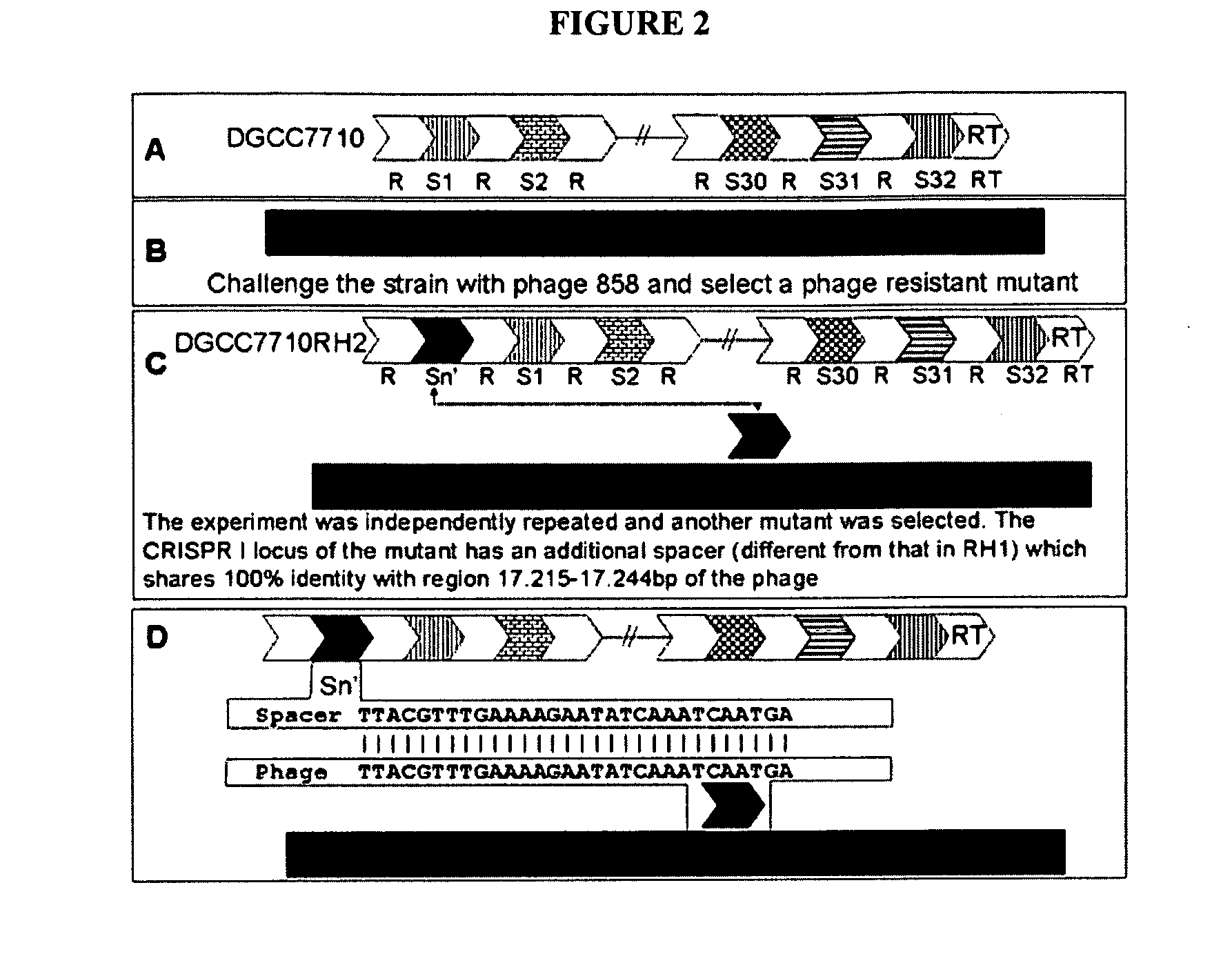
[0500]In these experiments, integration of a CRISPR spacer into the CRISPR locus was shown to provide resistance against a bacteriophage to which the CRISPR spacer shows identity. In these experiments, S. thermophilus strain DGCC7710RH1 was produced.
[0501]Streptococcus thermophilus strain DGCC7710 (deposited at the French “Collection Nationale de Cultures de Microorganismes” under number CNCM I-2423) possesses at least 3 CRISPR loci: CRISPR1, CRISPR2, and CRISPR3. In S. thermophilus strains CNRZ1066 and LMGI8311 for which the complete genome sequence is known (Bolotin et al., Microbiol., 151:2551-1561 [2005]. CRISPR1 is located at the same chromosomal locus: between str0660 (or stu0660) and str0661 (or stu0661).
[0502]In strain DGCC7710, CRISPR1 is also located at the same chromosomal locus, between highly similar genes. CRISPR1 of strain DGCC7710 contains 33 repeats (including the terminal repeat), and thus 32 spacers.
[0503]All these spacers are differ...
example 3
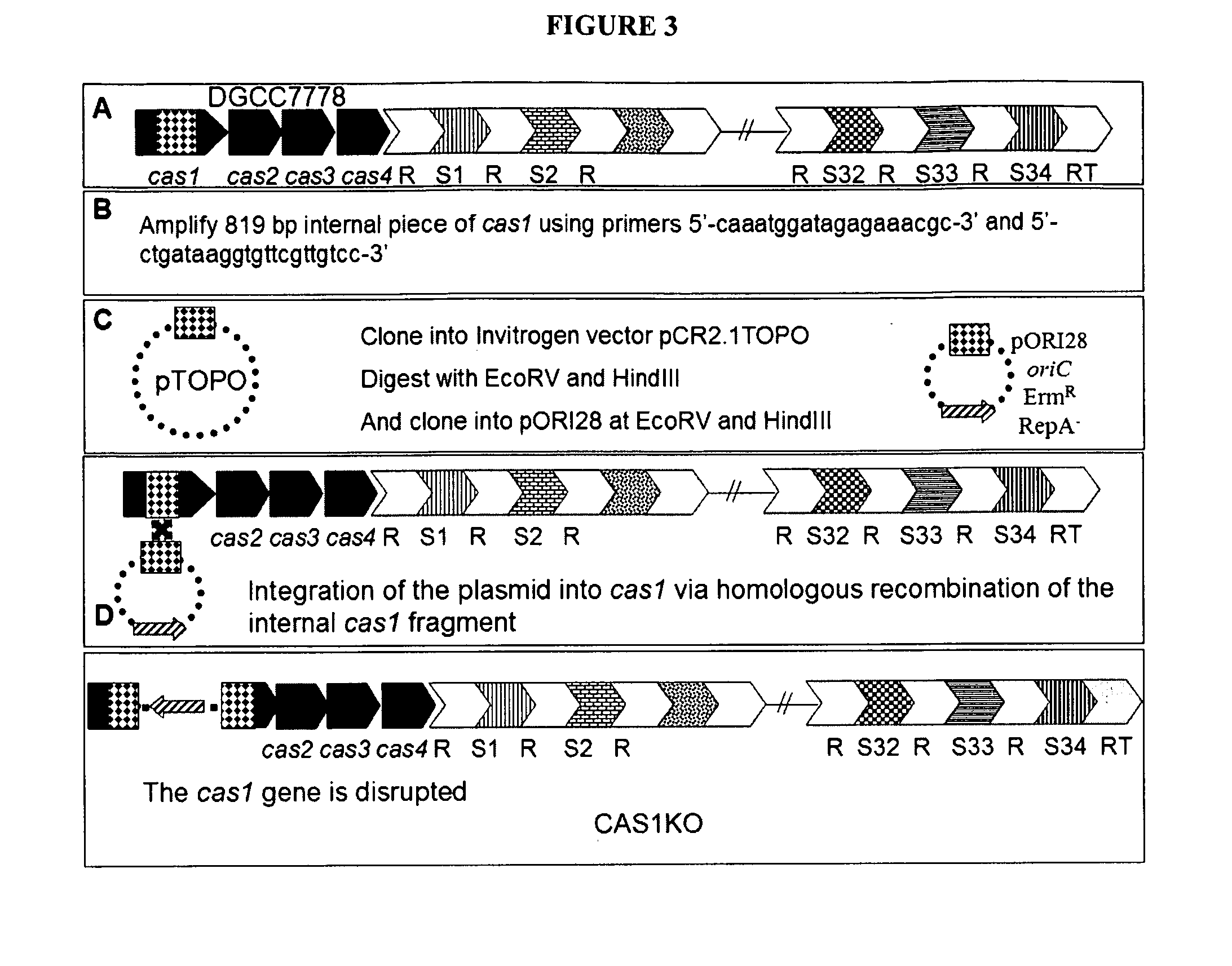
Construct Integration and Knockout
[0524]In this Example, methods used for construct integration and knockout are described.
[0525]The strains used in these experiments were:
S. thermophilus DGCC7710 parent strain, sensitive to phages 858 and 2972
S. thermophilus DGCC7778 CRISPR mutant resistant to 858
S. thermophilus DGCC7778cas1KO
S. thermophilus DGCC7778cas4KO
S. thermophilus DGCC7778RT
S. thermophilus DGCC7778RT′
S. thermophilus DGCC7710R2CRISPR mutant resistant to 2972
S. thermophilus DGCC7710R2S1S2
E. coli EC1,000 provided pOR128 (See, Russell and Klaenhammer, Appl. Environ. Microbiol., 67:43691-4364 [2001])
Escherichia coli pCR2.1TOPO provided pTOPO (See, Invitrogen catalog #K4500-01)
[0526]The following plasmids were used in these experiments:
pTOPO, a plasmid used for sub-cloning of the various constructs
pTOPOcas1ko contains an integral fragment of cas1
pTOPOcas4ko contains an integral fragment of cas4
pTOPOS1S2 contains the S1S2 spacer construct
pTOPO RT contains the RT terminal repeat con...
PUM
| Property | Measurement | Unit |
|---|---|---|
| or more nucleic acid | aaaaa | aaaaa |
| selective pressure | aaaaa | aaaaa |
| nucleic acid | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com