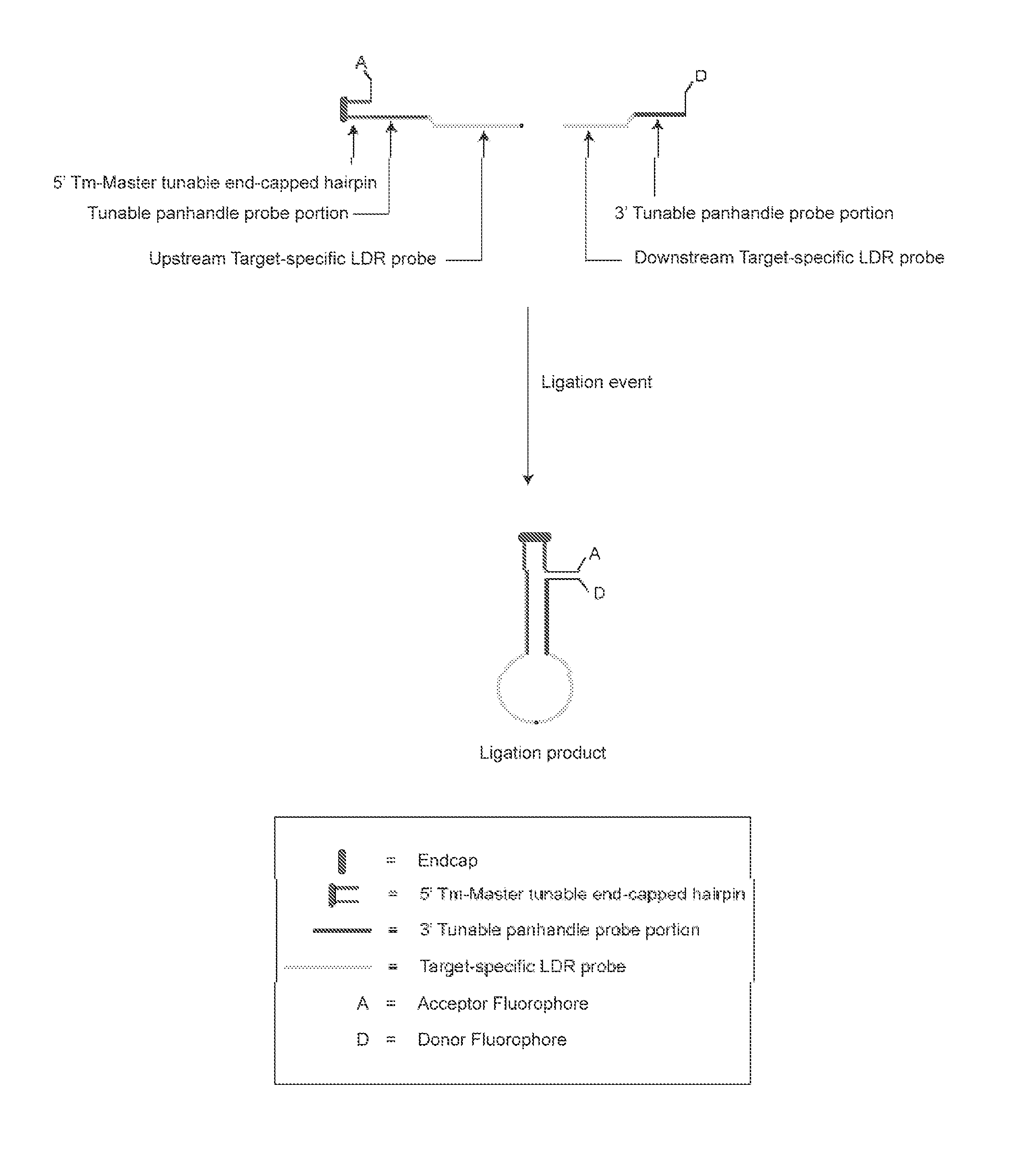Detection of target nucleic acid sequences using fluorescence resonance energy transfer
a technology of fluorescence resonance and target nucleic acid, which is applied in the field of detection of target nucleic acid sequences using fluorescence resonance energy transfer, can solve the problems of difficult design of ldr probes having the same acceptor, lack of multiplexing capability, and inability to obtain multiplexed readout signals, etc., to overcome the deficiencies of spfret-ldr probes, easy to distinguish, and tight melting profile
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
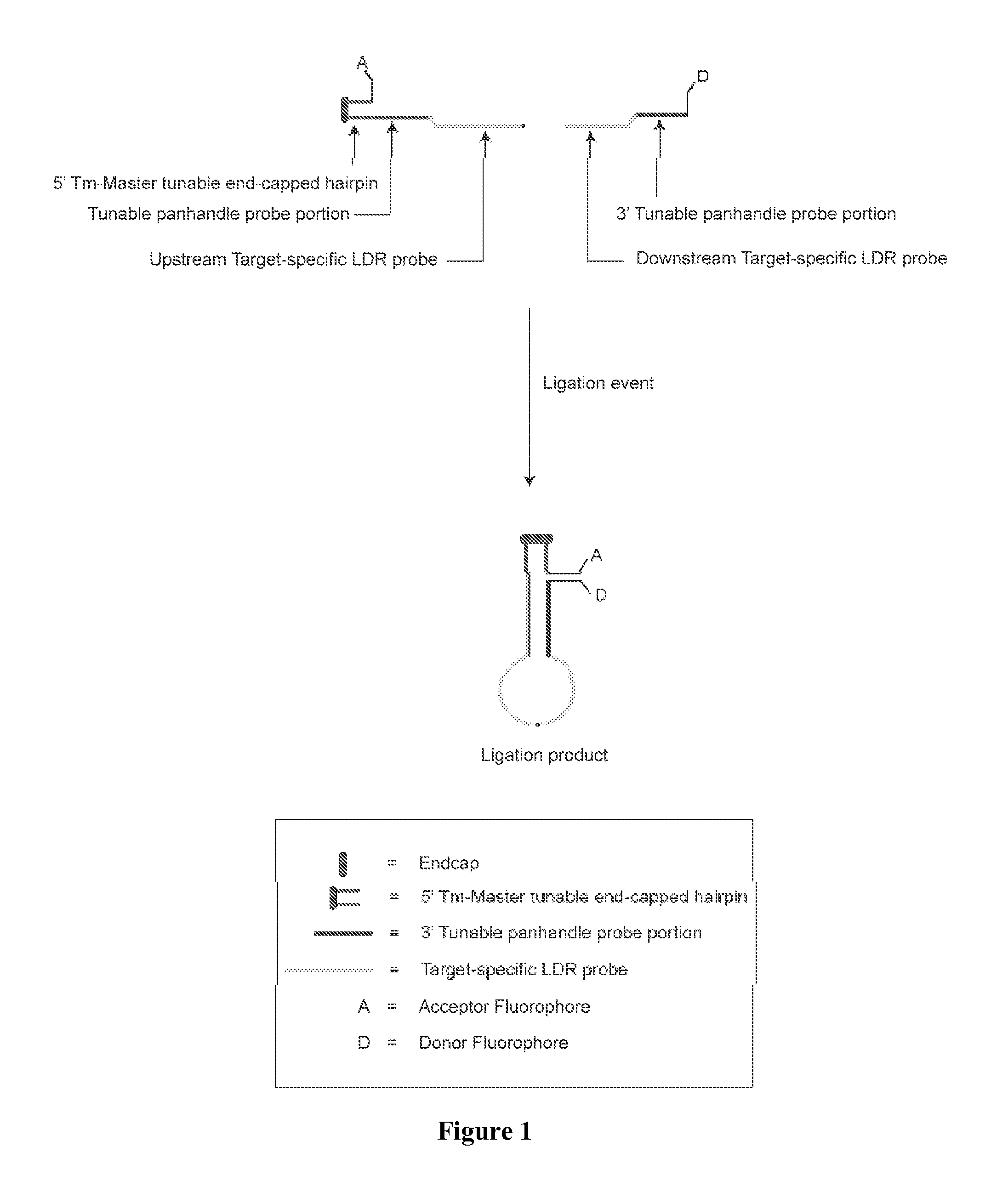
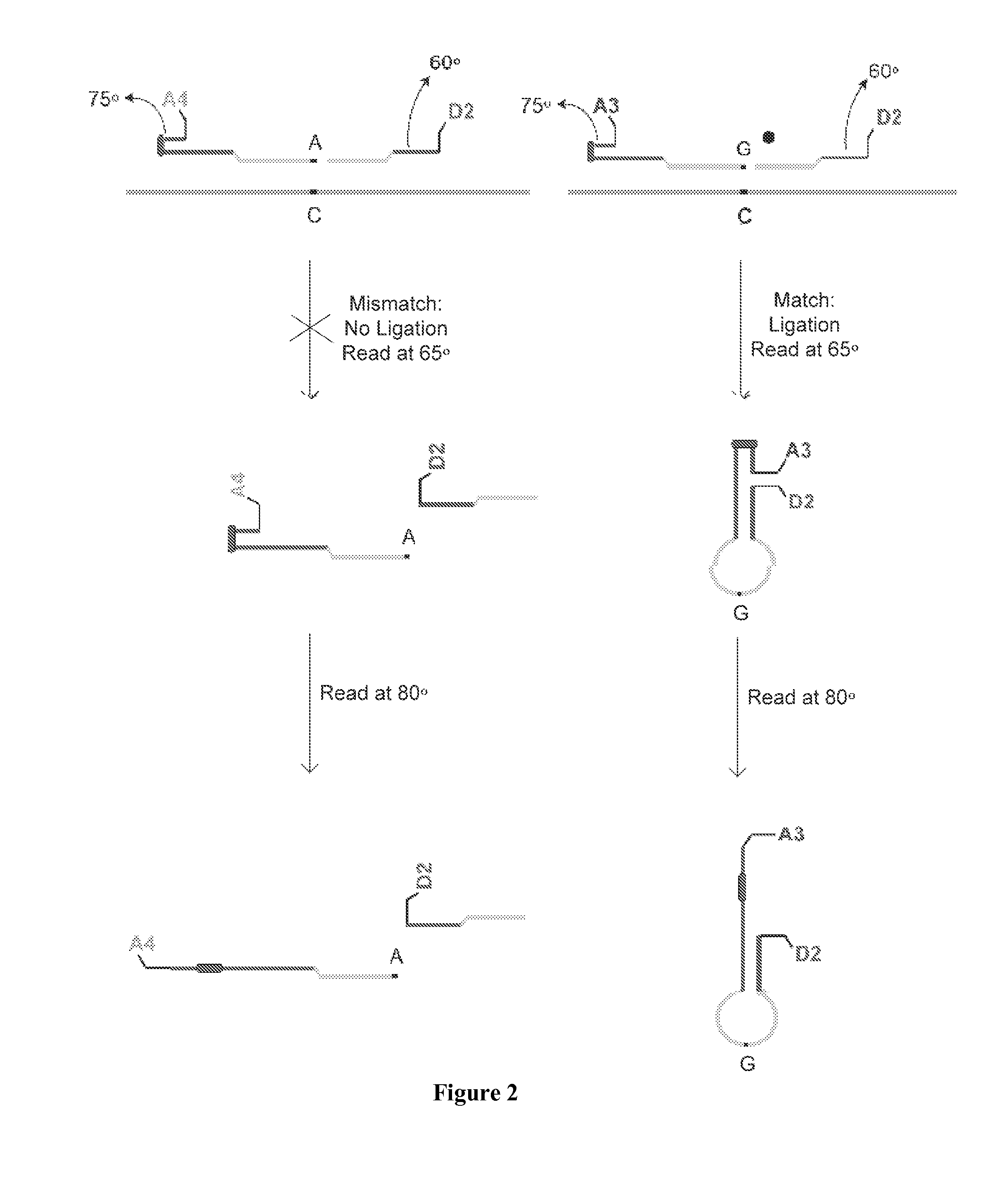
[0058]The present invention is directed to a method for identifying one or more of a plurality of target nucleic acid molecules in a sample. This method includes providing a sample potentially containing one or more target nucleic acid molecules and a plurality of oligonucleotide probe sets. Each probe set is characterized by (a) a first oligonucleotide probe, having a target-specific portion and a tunable portion with an endcapped hairpin and (b) a second oligonucleotide probe having a target specific portion and a tunable portion, wherein one of the first and second oligonucleotide probes has an acceptor group and the other of the first and second probes has a donor group. A ligase is provided and blended with the sample and the plurality of oligonucleotide probe sets to form a ligase detection reaction mixture. The mixture is subjected to one or more ligase detection reaction cycles with each cycle comprising a denaturation and hybridization treatment. During the denaturation tre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting temperatures | aaaaa | aaaaa |
| melting temperatures | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com