High throughput detection of genomic copy number variations
a technology of copy number variation and detection method, which is applied in the direction of material testing goods, biochemistry apparatus and processes, instruments, etc., can solve the problems of inability to identify the exact change in sequence with this method, disadvantages of on-chip extension/amplification methods compared to differential hybridization based methods, and the need for amplification and purification reactions, etc., to achieve high specificity and sensitivity. sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
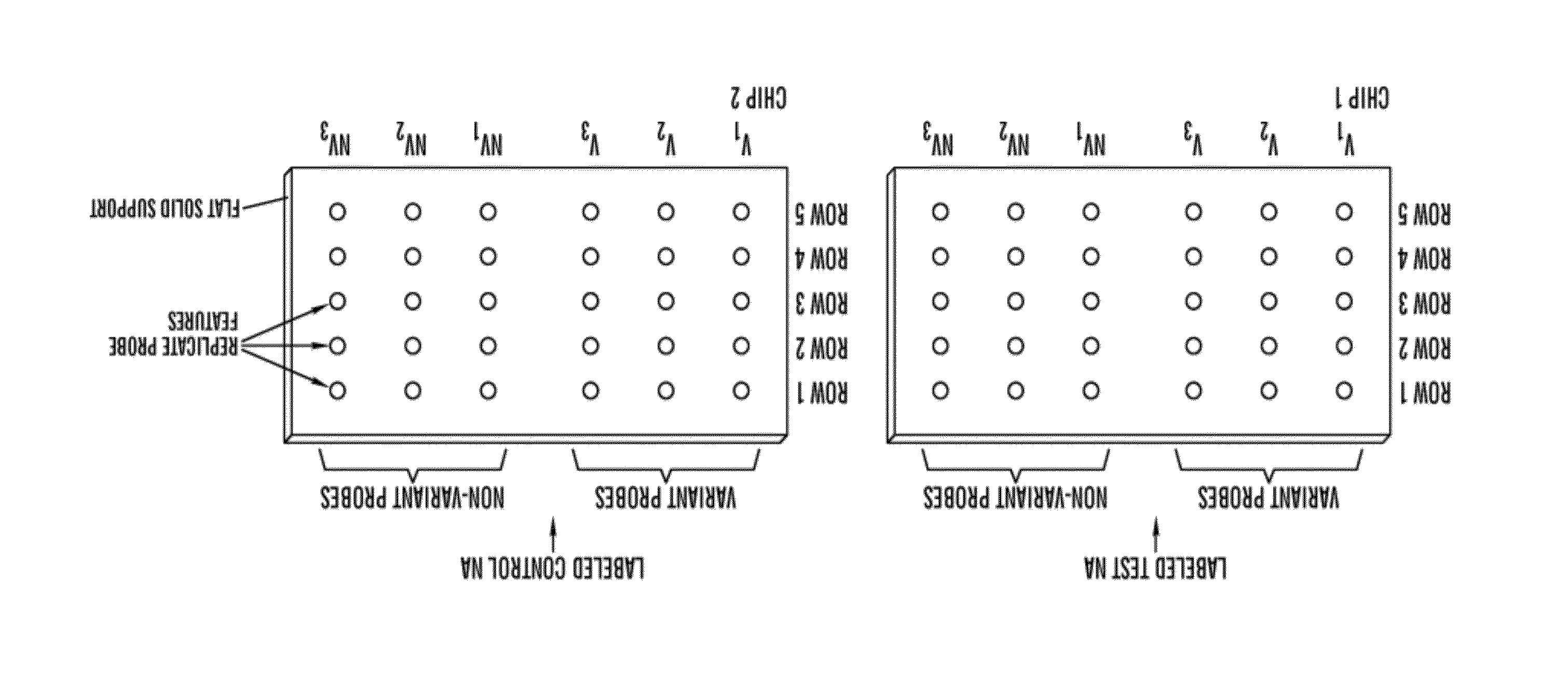
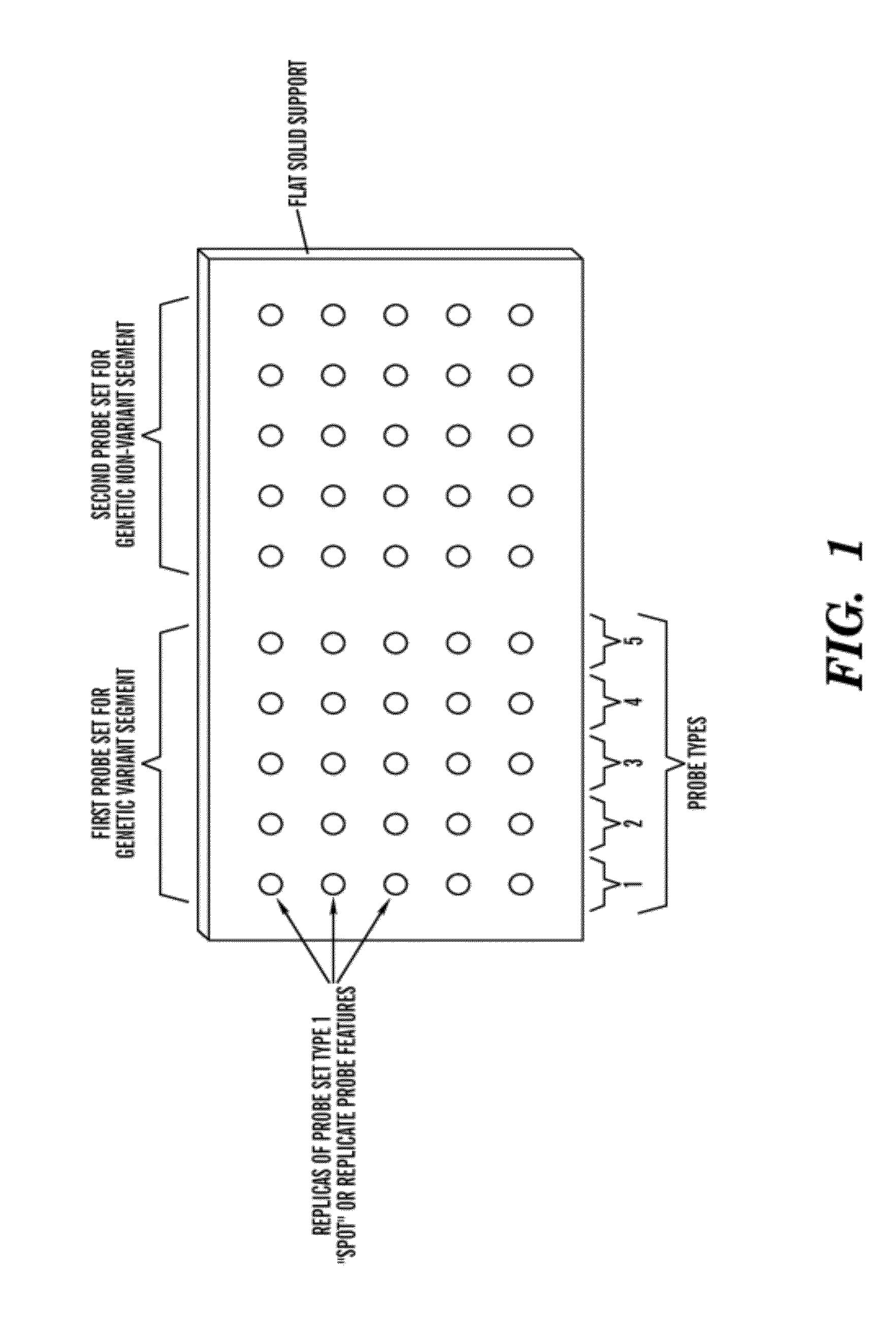
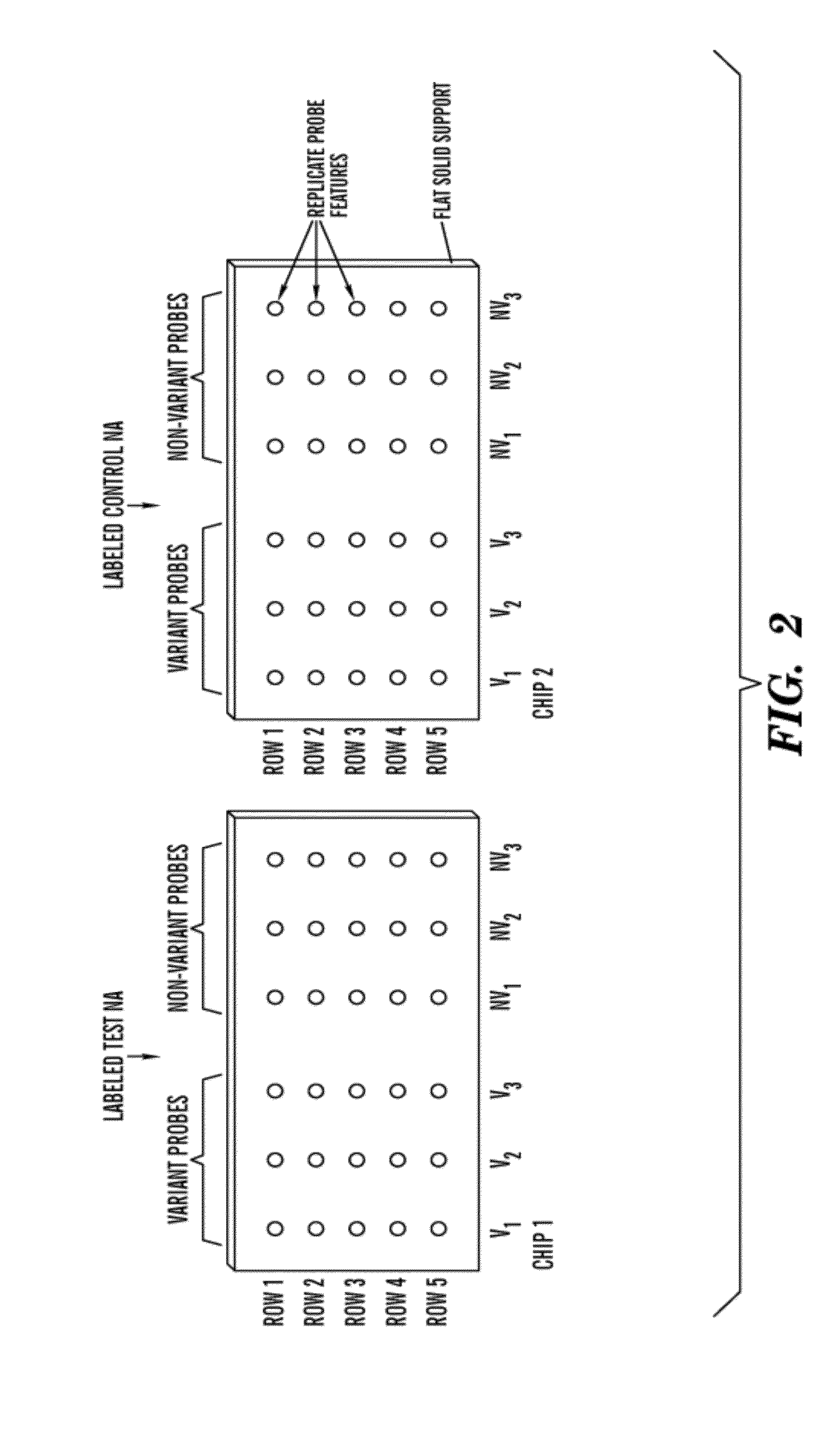
[0028]The present invention relates to an in vitro method of detecting genetic variations in an individual, specifically variations (e.g. duplications, multiple copies, deletion or loss of copies) in sequence segments in the genome. The inventors have developed a sensitive, specific and reproducible computer implemented method for simultaneously detecting and characterizing genetic variations in a genome. The inventors also developed methods for designing oligonucleotides used for carrying out the method of detecting genetic variations. Specifically, using the analysis methods described herein, one can analyze copy number variations (CNV) in a genome. The method is also useful for the development of products for genotyping these CNV. For example, in one embodiment, one can perform the method of genotyping according to the methods of the present invention to detect deletions of 1, 2 or 3 bases on AFFYMETRIX® re-sequencing chips.
[0029]The method is unique in that it is based on a comb...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
| densities | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com