I-crei meganuclease variants with modified specificity, method of preparation and uses thereof
a meganuclease and specificity technology, applied in the field of i-crei meganuclease variants with modified specificity, method of preparation and use thereof, can solve the problems of low efficiency, limited technology, and inability to have a natural meganuclease cleaving a gene of interes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Screening for New Functional Endonucleases
[0141]The method for producing meganuclease variants and the assays based on cleavage-induced recombination in mammal or yeast cells, which are used for screening variants with altered specificity, are described in the International PCT Application WO 2004 / 067736. These assays result in a functional LacZ reporter gene which can be monitored by standard methods (FIG. 3a).
A) Material and Methods
a) Construction of Mutant Libraries
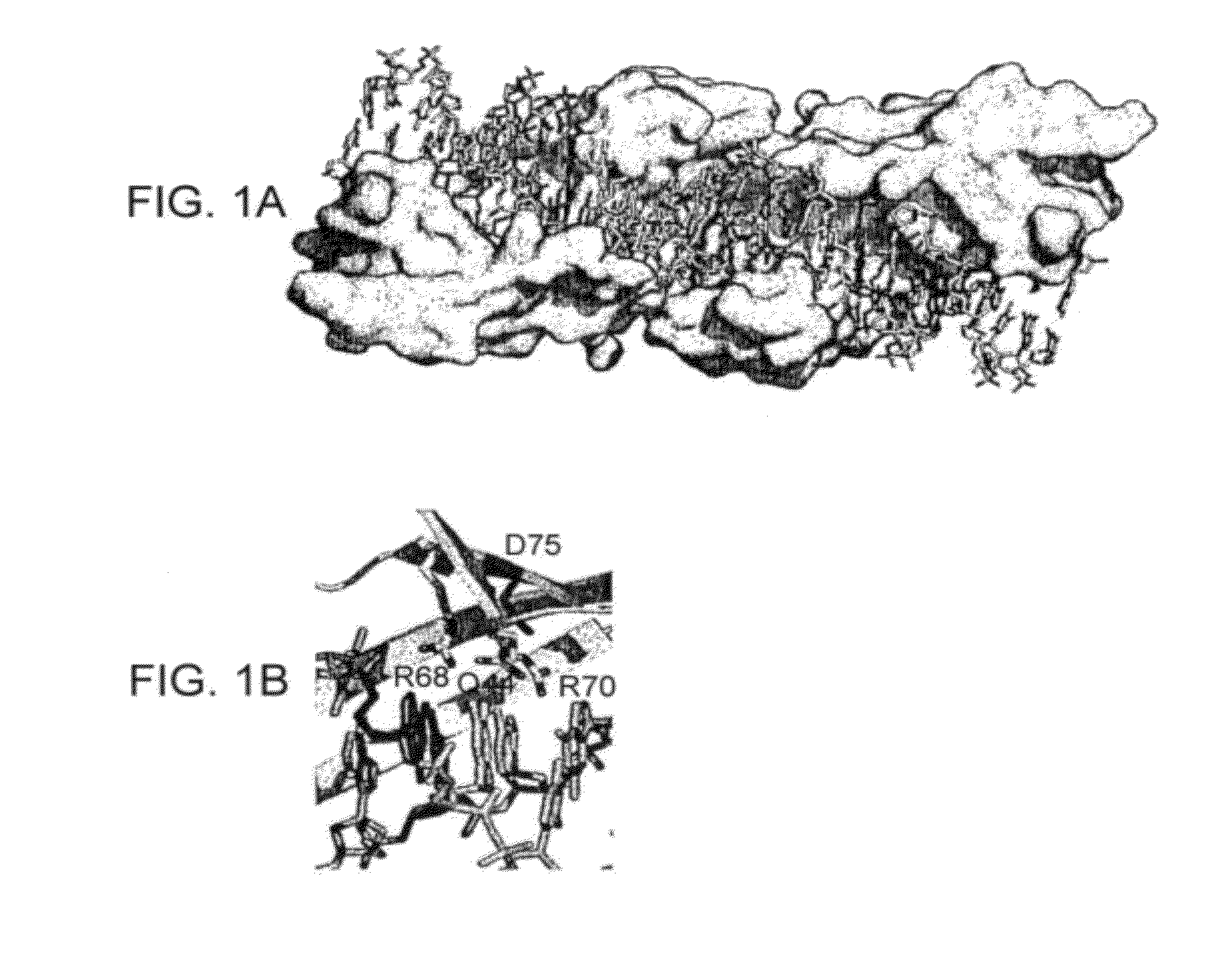
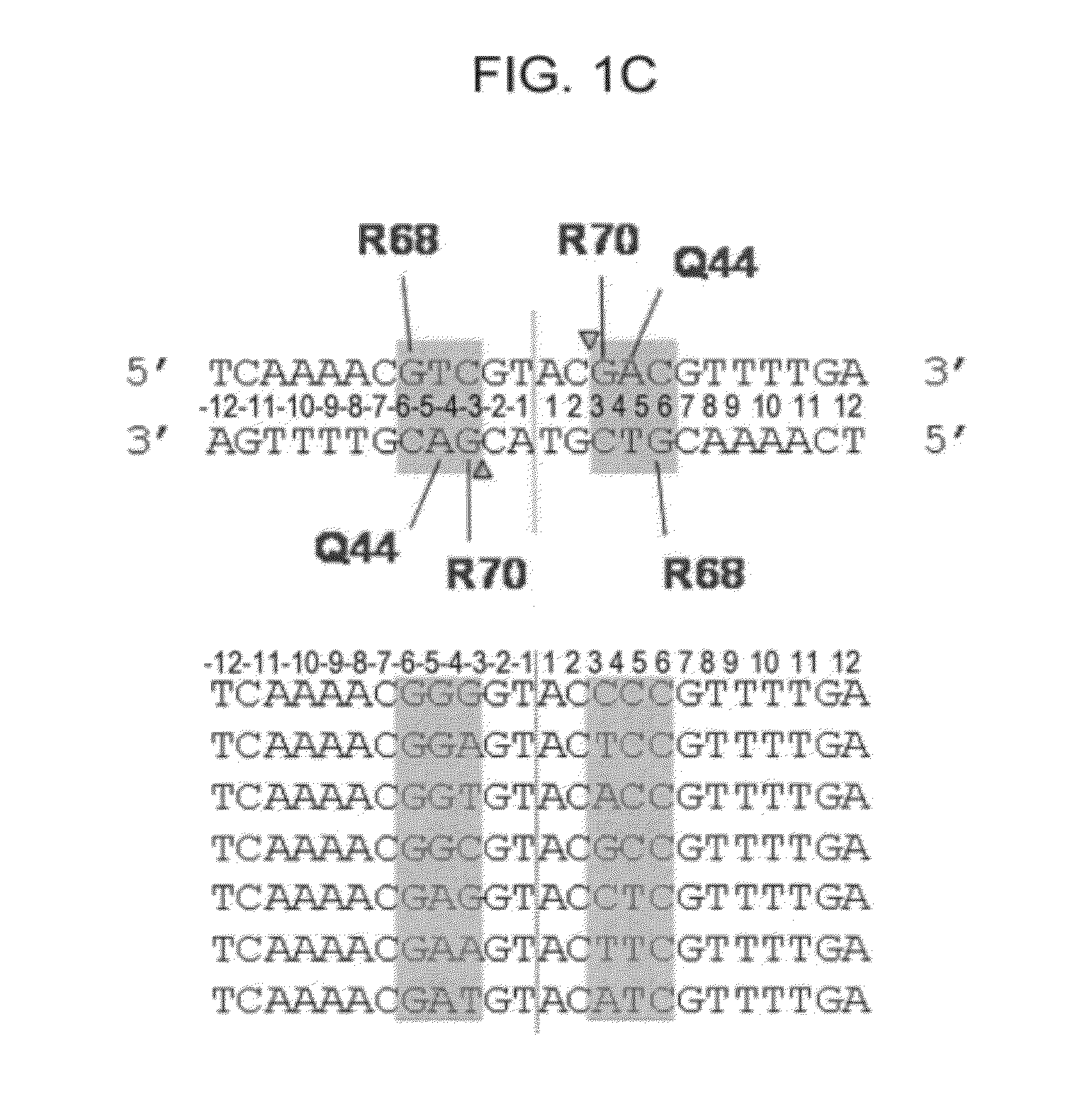
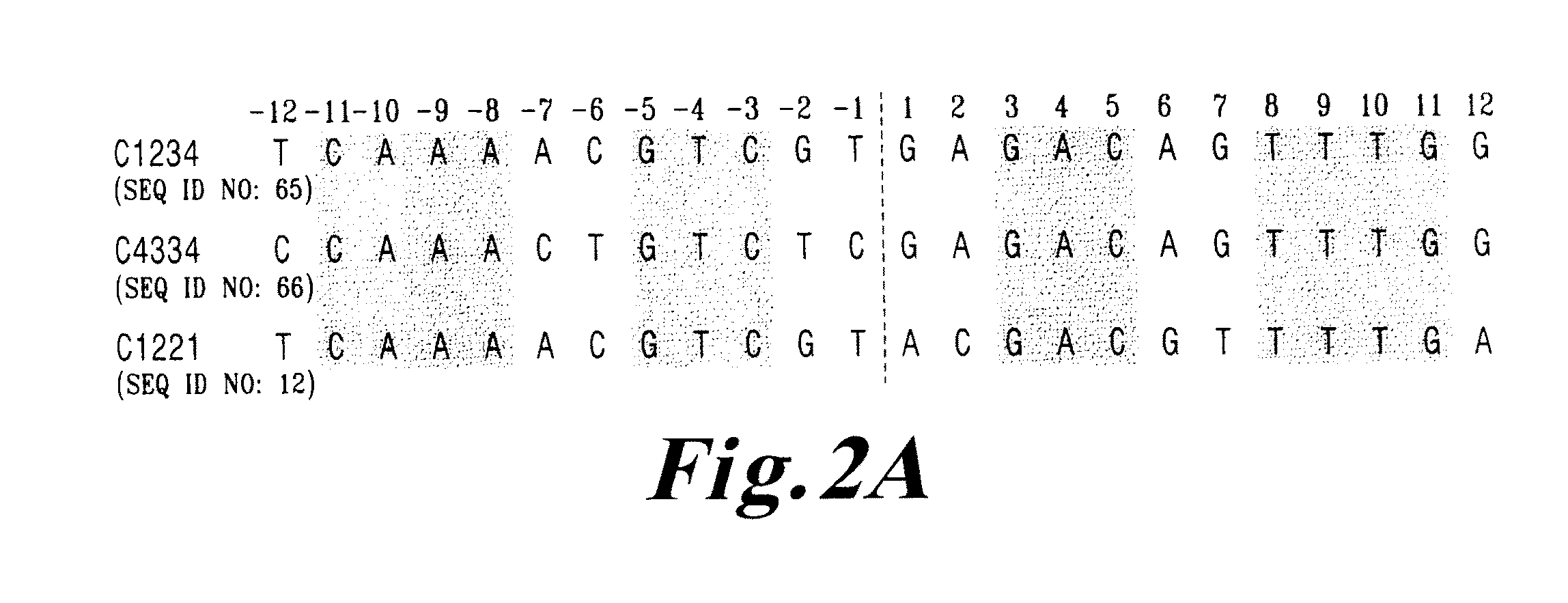
[0142]I-CreI wt and I-CreI D75N (or I-CreI N75) open reading frames (SEQ ID NO:69, FIG. 4A) were synthesized, as described previously (Epinat et al., N.A.R., 2003, 31, 2952-2962). Mutation D75N was introduced by replacing codon 75 with aac. The diversity of the meganuclease library was generated by PCR using degenerate primers from Sigma harboring codon VVK (18 codons, amino acids ADEGHKNPQRST) at position 44, 68 and 70 which interact directly with the bases at positions 3 to 5, and as DNA template, the I-CreI D75N gen...
example 2
I-CreI Meganuclease Variants with Different Cleavage Profiles
[0153]The validated clones from example 1 showed very diverse patterns. Some of these new profiles shared some similarity with the initial scaffold whereas many others were totally different. Various examples of profiles, including wild-type I-CreI and I-CreI N75, are shown in FIGS. 8 and 9a. The overall results (only for the 292 variants with modified specificity) are summarized in FIG. 7.
[0154]Homing endonucleases can usually accommodate some degeneracy in their target sequences, and one of our first findings was that the original I-CreI protein itself cleaves seven different targets in yeast. Many of our mutants followed this rule as well, with the number of cleaved sequences ranging from 1 to 21 with an average of 5.0 sequences cleaved (standard deviation=3.6). Interestingly, in 50 mutants (14%), specificity was altered so that they cleaved exactly one target. 37 (11%) cleaved 2 targets, 61 (17%) cleaved 3 targets and ...
example 3
Novel Meganucleases can Cleave Novel Targets While Keeping High Activity and Narrow Specificity
A) Material and Methods
a) Construction of Target Clones
[0156]The 64 palindromic targets were cloned into pGEM-T Easy (PROMEGA), as described in example 1. Next, a 400 bp PvuII fragment was excised and cloned into the mammalian vector pcDNA3.1-LACURAZ-ΔURA, described previously (Epinat et al., precited). The 75 hybrid targets sequences were cloned as follows: oligonucleotides were designed that contained two different half sites of each mutant palindrome (PROLIGO).
b) Re-Cloning of Primary Hits
[0157]The open reading frame (ORF) of positive clones identified during the primary screening in yeast was recloned in: (i) a CHO gateway expression vector pCDNA6.2, following the instructions of the supplier (INVITROGEN), and ii) a pET 24d(+) vector from NOVAGEN Resulting clones were verified by sequencing (MILLEGEN).
c) Mammalian Cells Assay
[0158]CHO-K1 cell line from the American Type Culture Collect...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular mass | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com