Process for multi-analyses of rare cells extracted or isolated from biological samples through filtration
a rare cell and multi-analysis technology, applied in the field of rare cell extraction or isolation from biological samples, can solve the problems of introducing selection bias, only recovering some rare cells in a sample, and not addressing the triple challenge of extracting or isolating rare cells, so as to reduce the relative number of rare cells, reduce the number of rate cells, and improve the effect of molecular structur
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
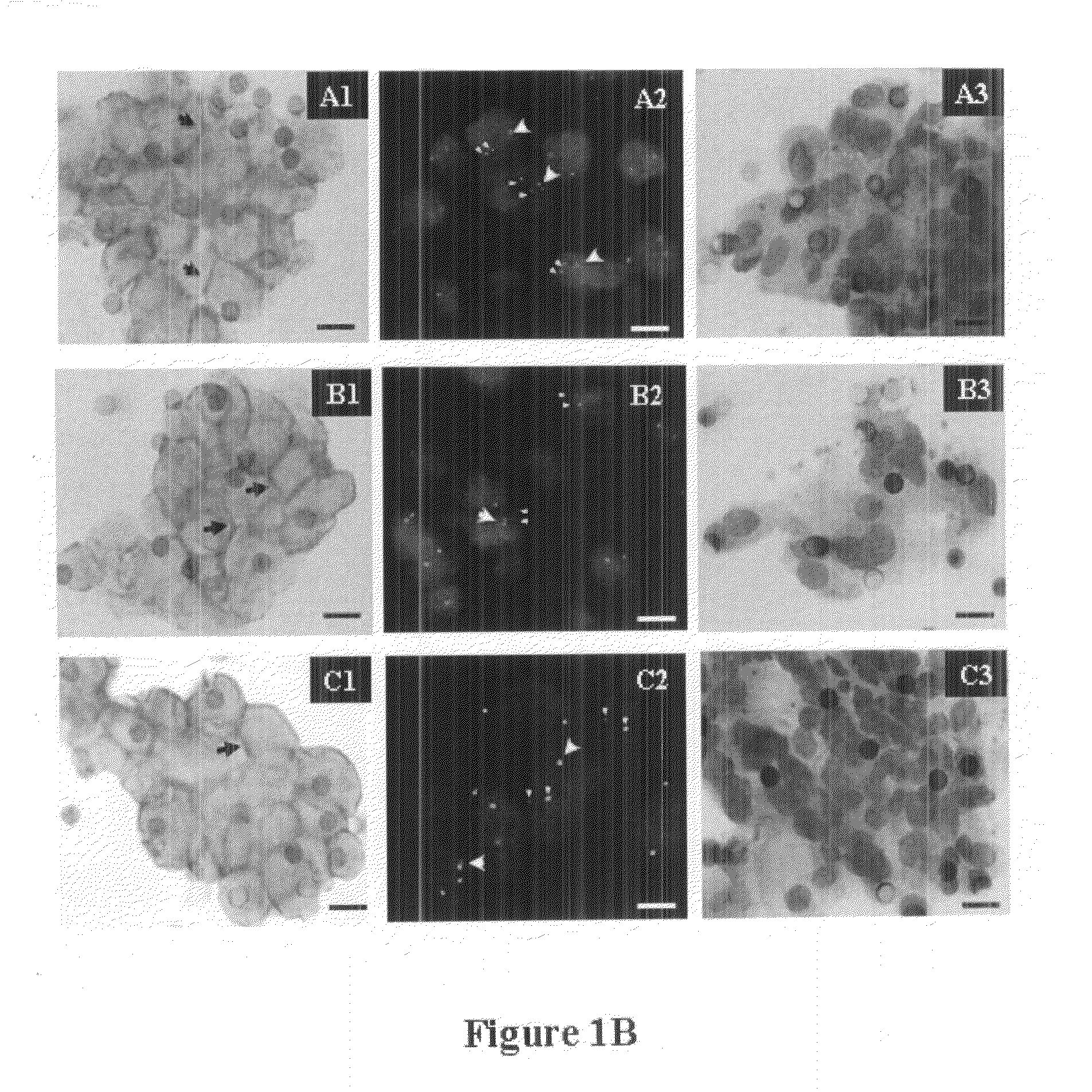
Multianalysis of Fresh, Non-Fixed, Tumor Cells Enriched from Blood by Filtration
[0202]Filtration.
[0203]Fresh tumor cells were extracted and enriched from blood by filtration as described in published patent application US-2009-0226957.
[0204]The extracted, non-fixed cells were stained to determine their morphology and their nucleic acids are extracted and analyzed by RT-PCR or PCR after whole genomic amplification.
[0205]To extract and enrich rare tumor cells from blood, a blood sample is diluted 20-fold using a buffer for red blood cell lysis. Lysis is allowed to proceed for 5 minutes at room temperature with a gentle agitation.
[0206]The treated blood sample is then immediately filtered at a depression of −6 mBar. Filtration is stopped before it is totally complete in a way that about 200 μL of solution remains in the well.
[0207]The enriched fresh tumor cell material is collected by gently pipetting 3 times 1 mL of cell culture media (DMEM HEPES 1% Fetal Calf Serum).
[0208]The collect...
example 2
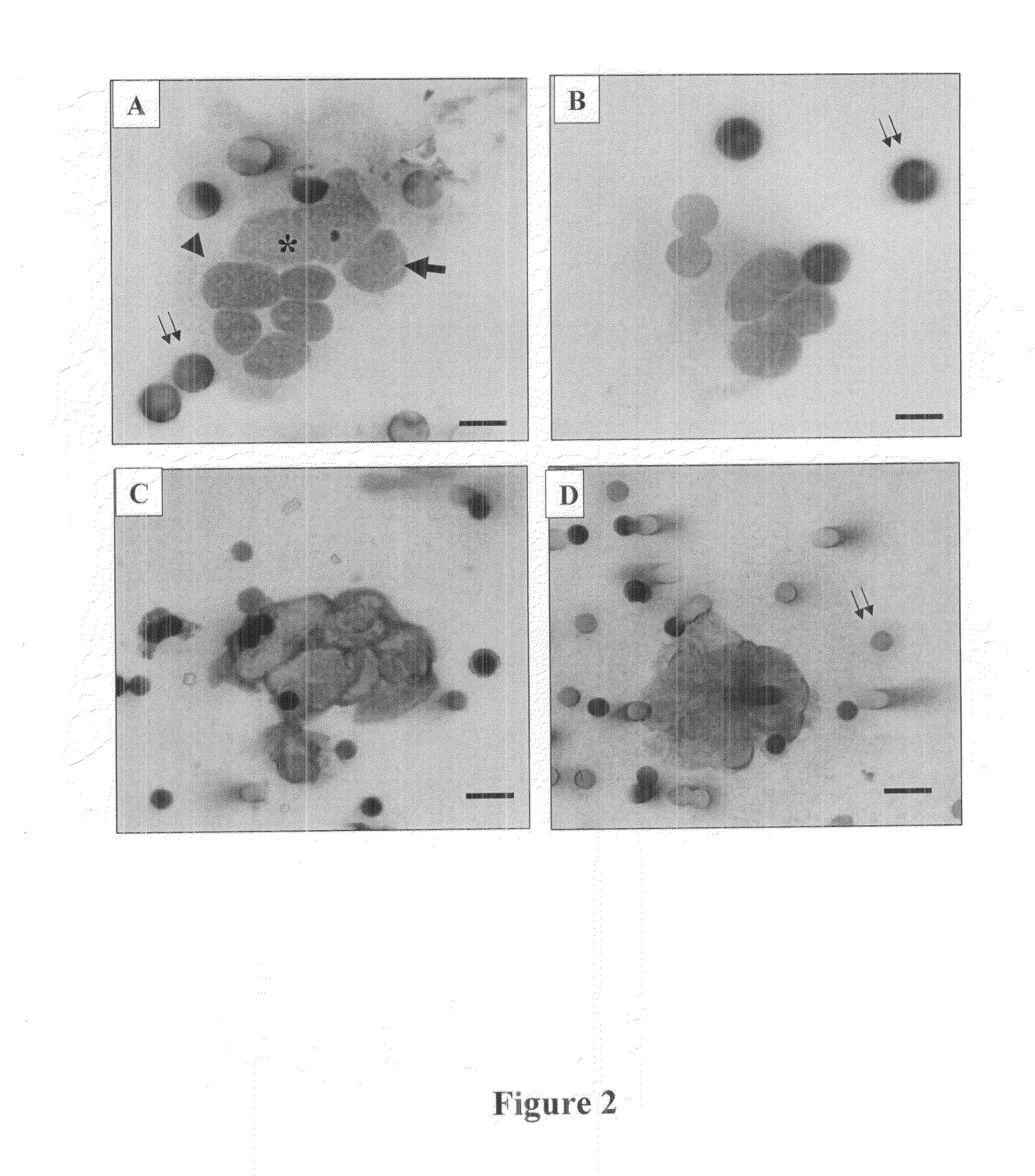
Genetic Characterization of Circulating Tumor Cells and Detection of a Theranostic Mutation in the Circulating Tumor Cells
[0234]According to a preferred mode of implementation, rare cells can be isolated from blood of patients with lung cancer by filtration, tumor cells can be identified among the isolated rare cells by cytomorphological analyses and characterized by ALK specific antibodies and molecular probes.
[0235]ALK-gene rearrangement, a comparative analysis on circulating tumor cells and tumor tissue from lung adenocarcinoma patients
[0236]The aim of this work described below as 1) to assess the ALK status in CTCs having malignant cytopathological criteria isolated by ISET in 87 patients with lung adenocarcinomas and, 2) to compare the ALK status found in CTCs and in the corresponding tumor tissue. For this purpose, an assay based on a dual immunochemical and FISH approach for ALK-gene rearrangement was used and applied to both CTCs and corresponding tumor tis...
example 3
Application of the Present Invention to Early Diagnosis of Invasive Cancers
[0263]“Sentinel” Circulating Tumor Cells allow early diagnosis of lung carcinoma in patients with chronic obstructive pulmonary disease.
[0264]Circulating tumor cells (CTC) are thought to circulate at a very early stage in invasive cancers; however, they have never been reported as invasive cancers first hallmark. We report about three out of 168 patients with Chronic Obstructive Pulmonary Disease (COPD), a pre-tumor condition of Non-Small Cell Lung Cancer (NSCLC), who were found to have CTC detected by ISET (Isolation by Size of Tumor Cells), a highly sensitive “cytopathology-based” CTC-platform. CT scan, performed yearly, detected a lung nodule only 1 to 4 years after CTC appeared in blood leading to its surgical resection and pathological diagnosis of early stage NSCLC. ISET was then repeated 9 or 12 months after surgery and showed disappearance of CTC. These 3 cases show, as a proof of concept, the potenti...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com