Antibody production
a technology of antibody and production, applied in the field of antibody production, can solve the problems of limited expression of transgenes across target tissues, non-productive response to antigen challenge, and technical demands, and achieve the effects of improving the probability of vh region, reducing the number of antigens, and increasing the diversity of antigens
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
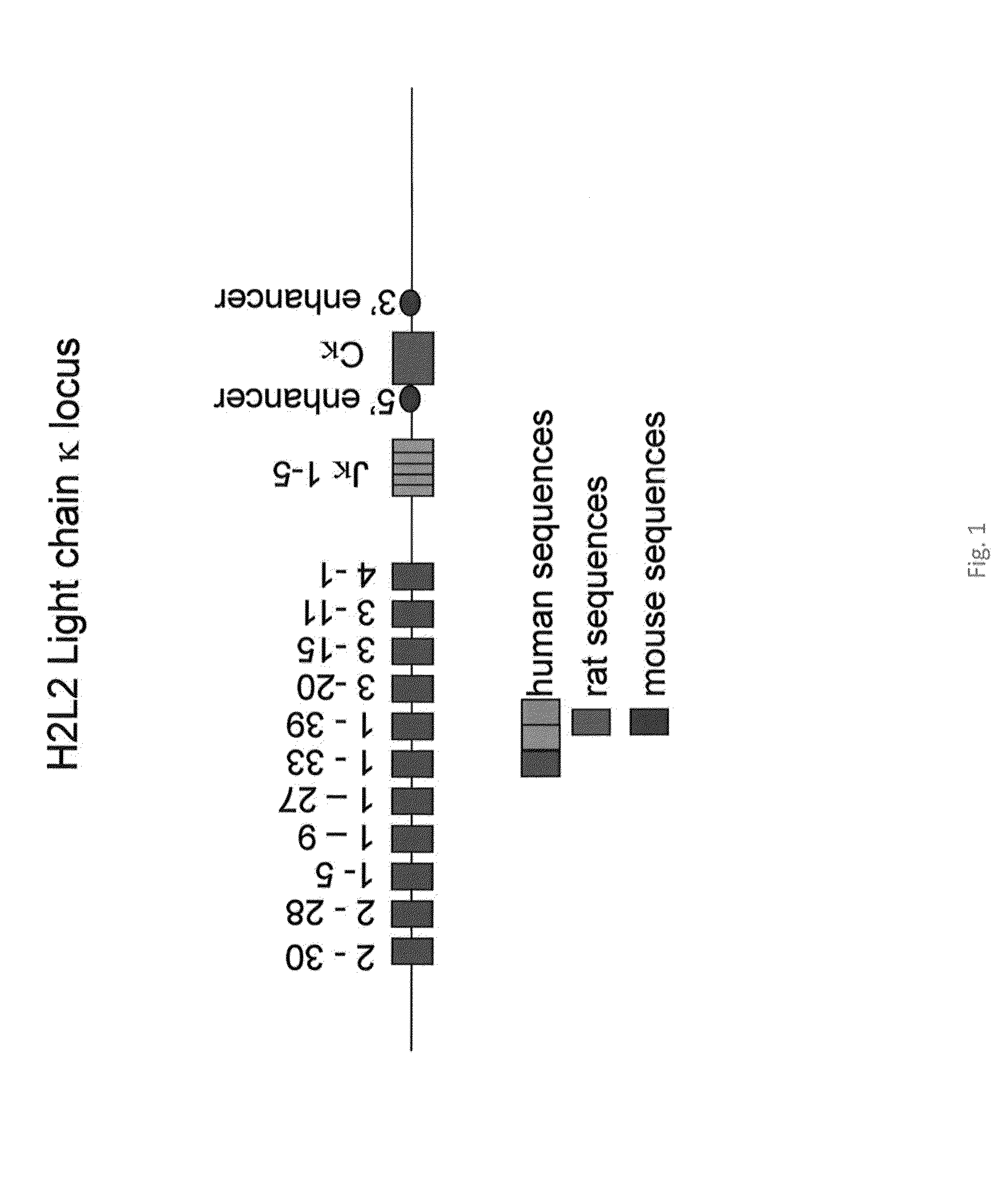
[0115]The most frequently and moderately frequently used Vκ genes of the human Igκ locus (FIG. 1, Vκ2-30, Vκ2-28, Vκ1-5, Vκ1-9, Vκ1-27, Vκ1-33, Vκ1-39, Vκ3-20, Vκ3-15, Vκ3-11, and Vκ4-1, assessed using the Ig database; http: / / imgt.cines.fr / ) were amplified by standard PCR and subcloned between XhoI / SalI sites, as described previously for human VH segments. This allows the multimerisation of the Vκ regions, keeping the multimer between XhoI and SalI sites.
[0116]Also, the 3′ end of the mouse κ locus, including the 3′κ enhancer, and the rat constant (Cκ) region plus the rat 5′ enhancer were cloned together. Next, the human Jκ region and the region (17 kb) from between mouse Vκ and Jκ were cloned in to maintain the normal spacing between Vκ regions and Jκ. Finally, the human Vκ were inserted into the PAC by routine procedures (e.g. Janssens et al. (2006), supra).
[0117]The Vκ segments were multimerized and ligated into the PAC vector containing the human J regions and the mouse enhancers...
example 2
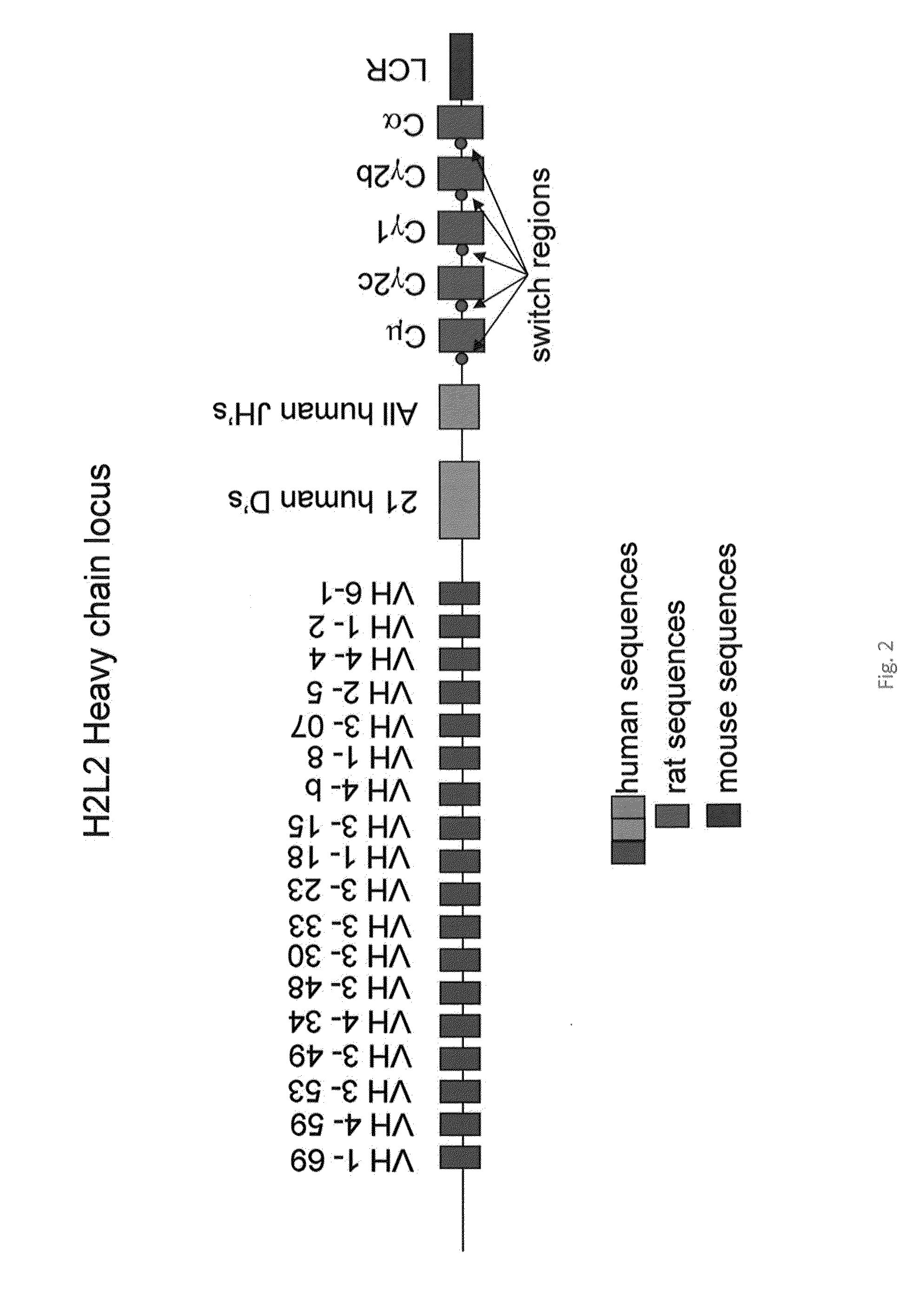
[0119]A hybrid human / rat IgH locus has been generated having 18 human VH segments and 5 rat constant regions (FIG. 2; human VH 6-1, VH 1-2, VH 4-4, VH 2-5, VH 3-07, VH 1-8, VH 4-b, VH 3-15, VH 1-18, VH 3-23, VH 3-33, VH 3-30, VH 3-48, VH 4-34, VH 3-49, VH 3-53, VH 4-59, VH 1-69). First, a central 70 kb DJ region of the human locus containing 21 D gene segments and all 6 J gene segments was extended at the 5′ end with 8 kb from the mouse IgH intron to maintain the proper distance between VH segments and the D region. Next the first VH region (6-1) of 10 kb with an artificial SceI meganuclease site was cloned at the 5′ end of the mouse intron sequences. In a separate plasmid, all the remaining VH regions were cloned together by slotting in Xho1 / SalI VH segments. The VH multimer was cloned into the VH6-1DJ plasmid, after which the rat constant regions were added to complete the locus (Cμ, Cγ2χ, Cγ1, Cγ2b, and Cα and switch regions). These have been amplified by standard long range PCR ...
example 3
[0121]The hybrid IgH and hybrid Igκ transgenic mice were subsequently bred to obtain mice that are positive for the human / rat hybrid IgH and Igκ expression. These mice were subsequently immunized to generate antigen-specific hybrid human / rat H2L2 antibodies by routine procedures.
PUM
| Property | Measurement | Unit |
|---|---|---|
| structure | aaaaa | aaaaa |
| affinity | aaaaa | aaaaa |
| color | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com