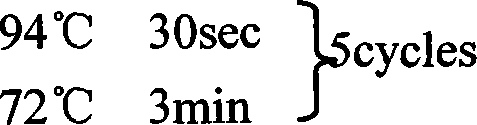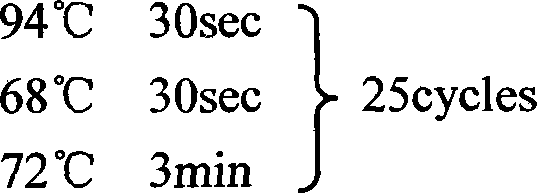DNA fragment related to pig polydactyly
A technology of fragments and peptides, which is applied in the field of mutation and genetic engineering, can solve the problems of no difference found and correlation analysis, etc., and achieve the effect of accurate detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0024] Example 1 Obtaining of DNA fragments of the present invention
[0025] 1 Collection of samples
[0026] Immediately after slaughtering pigs, collect heart, liver, spleen, lung, kidney, forelimb muscles and other tissues, cut into small pieces, put them in cryopreservation tubes, store them in liquid nitrogen immediately, and place them in a -80°C ultra-low temperature freezer after returning to the laboratory save in .
[0027] 2 Searching for porcine homologous EST and designing primers
[0028] Taking the coding sequence of the human / mouse polydactyly candidate gene C7orf2 / Lmbr1 gene as the search object, BLAST was performed on the NCBI website to obtain a pig homologous EST (CV878120, 724bp), and the 6bp-636bp part of the sequence is similar to the human C7orf2 The homology of the 497bp-1148bp part of the gene sequence ((accession number: NM_022458, 2781bp in total, 155bp-1627bp part is CDS)) is 88%. Based on this EST sequence, use Primer5.0 software to design pri...
example 2
[0149] Example 2 Molecular detection of pig developmental traits
[0150] (1) extraction of pig blood total RNA;
[0151] Collect 5 ml of jugular venous blood from individual pigs to be tested, and anticoagulate with heparin. After separating leukocytes, use the total RNA extraction reagent (D312) of Dalian Bao Bioengineering Co., Ltd. to total RNA according to the instructions.
[0152] (2) RT-PCR amplification;
[0153] Use SMARTTM RACE cDNA Amplification kit, reverse transcription reaction: total RNA 1-3ul, 3'-CDSprimer 1ul, add deionized water to 5ul, 70°C for 2min, put on ice for 2min, add 5×First-Strand Buffer 2ul, DTT (20mM) 1ul, dNTP (10mM) 1ul, BD PowerScript Reverse Transcriptase 1ul, 90min at 42°C, diluted with 100ul Tricine-EDTA Buffer, bathed in water at 72°C for 7min, to obtain 3'-RACE-Ready cDNA.
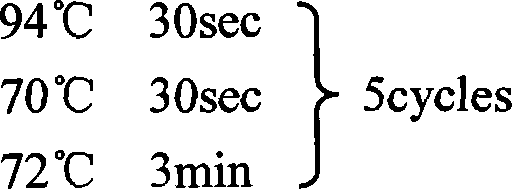
[0154] The first round of PCR was performed using UPM (provided in the kit) and GSP2 (5'-GAC TTG CCG CTC ATT GAC AAC GAC-3'). See the kit instructions for the PCR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com