An L-amino acid-producing bacterium and a method for producing an L-amino acid
An amino acid, bacterial technology, applied in the field of L-threonine and L-tryptophan to produce L-amino acids such as L-lysine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
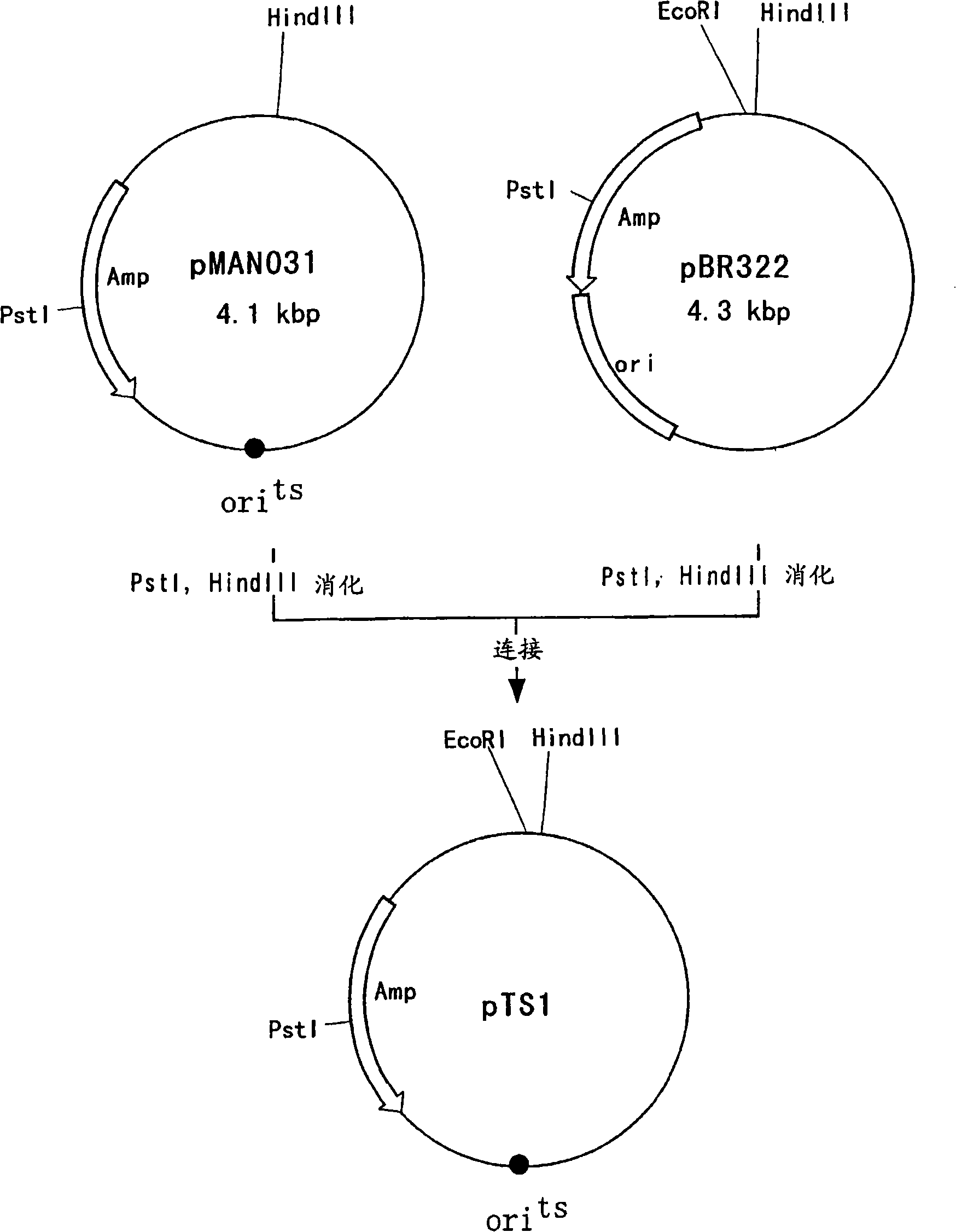
[0155] Embodiment 1: construction is used for enhancing the plasmid of evgAS, gadE and ydeO gene
[0156] 1-1. Construction of plasmids for enhancing evgA and evgS genes
[0157] The complete nucleotide sequence of the genome of E. coli (E. coli K-12 strain) has been reported (Science, 277, 1453-1474 (1997)), and evgAS is known to form an operon structure. Based on the nucleotide sequence of the evgAS gene reported in the above report (GenBank accession numbers AAC75428 and AAC75429), an oligonucleotide shown in SEQ ID NO: 17 having a HindIII site was synthesized as a 5' primer, and an oligonucleotide having an EcoRI site The oligonucleotide shown in SEQ ID NO: 18 of the spot served as the 3' primer. Using these primers, PCR was performed using the genomic DNA of Escherichia coli W3110 strain as a template, and the amplified product was treated with restriction endonucleases HindIII and EcoRI to obtain a fragment containing the evgAS gene. The purified PCR product was ligate...
Embodiment 2
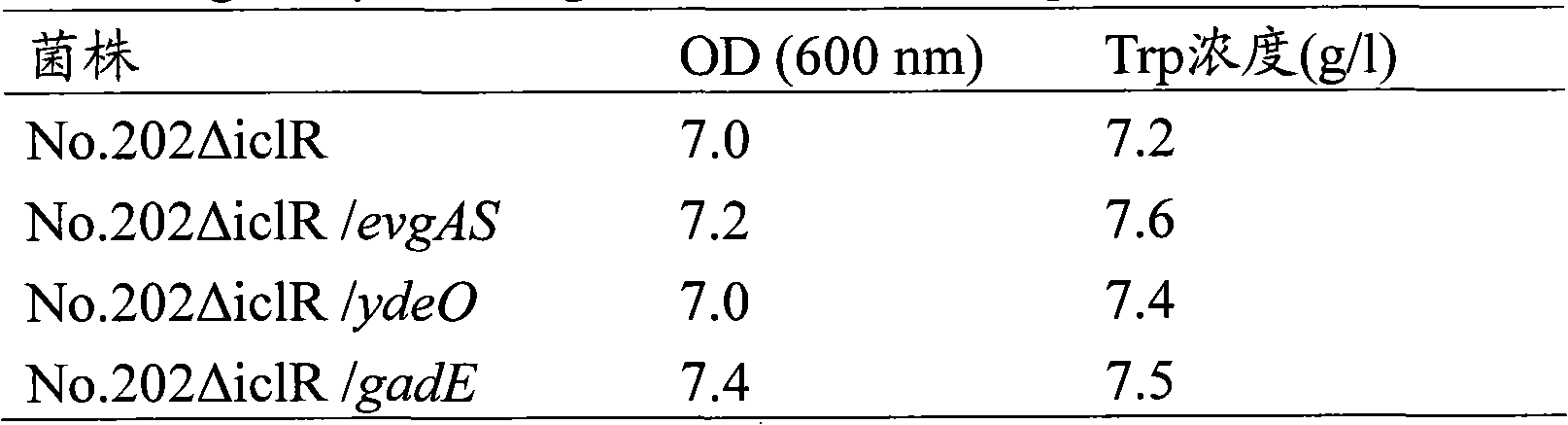
[0162] Example 2: Effects of evgAS, ydeO and gadE gene amplification on L-tryptophan-producing bacterial strains of the genus Escherichia
[0163] The L-tryptophan-producing strain No. .202ΔiclR, thereby obtaining ampicillin-resistant strains. After confirming that these plasmids have been introduced, the bacterial strain introduced with the gadE-amplified plasmid pMW-gadE was named No.202ΔiclR / gadE strain; the bacterial strain introduced with the evgAS-amplified plasmid pMW-evgAS was named No.202ΔiclR / evgAS strain; And the strain into which the ydeO-amplified plasmid pMW-ydeO was introduced was named No. 202ΔiclR / ydeO strain.
[0164] The evgAS, ydeO, and gadE genes can be amplified in the same manner using known L-tryptophan-producing bacteria other than No. 202ΔiclR.
[0165] The strain produced as above was cultured in LB medium containing 50 mg / L ampicillin at 37°C until OD600 became about 0.6. Thereafter, an equal volume of 40% glycerol solution was added to the cultu...
Embodiment 3
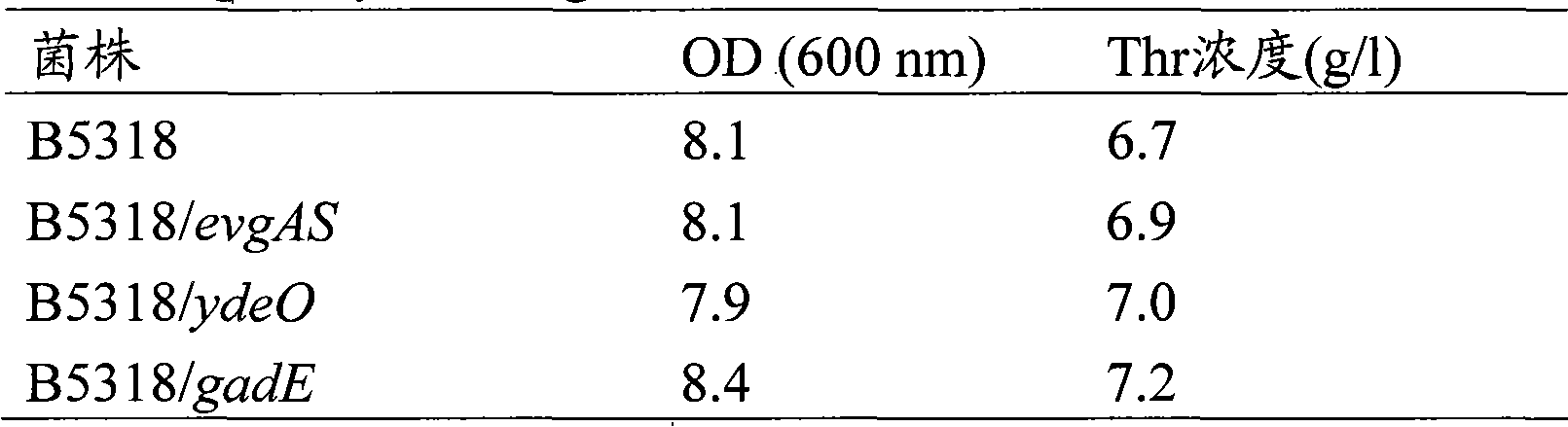
[0186] Example 3: Effect of evgAS, ydeO and gadE amplification on L-threonine-producing bacterial strains of the genus Escherichia
[0187] Escherichia coli VKPM B-5318 strain (see EP0593792) was used as the L-threonine-producing bacterial strain of the genus Escherichia.
[0188] The VKPM B-5318 strain was transformed with the gadE-amplification plasmid pMW-gadE, evgAS-amplification plasmid pMW-evgAS and ydeO-amplification plasmid pMW-ydeO constructed in Example 1, thereby obtaining an ampicillin-resistant strain. After confirming that these plasmids have been introduced, the strain into which the gadE-amplification plasmid pMW-gadE was introduced was named B5318 / gadE strain; the strain into which evgAS-amplification plasmid pMW-evgAS was introduced was named B5318 / evgAS strain; and ydeO was introduced -The strain amplifying the plasmid pMW-ydeO was named B5318 / ydeO strain.
[0189] The evgAS, ydeO, and gadE genes can be amplified in the same manner using known L-threonine-p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com