Method of elevating GGT activity of plant, plant with elevated GGT activity and method of constructing the same
A plant, a technology of transgenic plants, applied to one or more of the group, the new utilization of GGT and its encoded gene, the field of gene utilization, can solve the problems of unreported disclosure, unknown genes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0084] Cultivation of all plants was carried out under the following conditions:
[0085] The following components were used on the plate as the minimal medium for culturing: PNS ( Mol. Gen. Genet. 204: 430-434) or MS (Physiol Plant 15: 473-479) inorganic salts and 0.8 (w / v) agar. If cultivation is carried out on rock wool, only PNS inorganic salts are used as a nutrient source.
[0086] In addition, the obtained Arabidopsis thaliana GGT deletion strain was used as a model plant for transformation experiments. The methods for obtaining GGT-deleted strains are shown in Reference Examples 1 and 2.
reference example 1
[0087] Reference Example 1. Obtaining a GGT-deleted strain of Arabidopsis thaliana
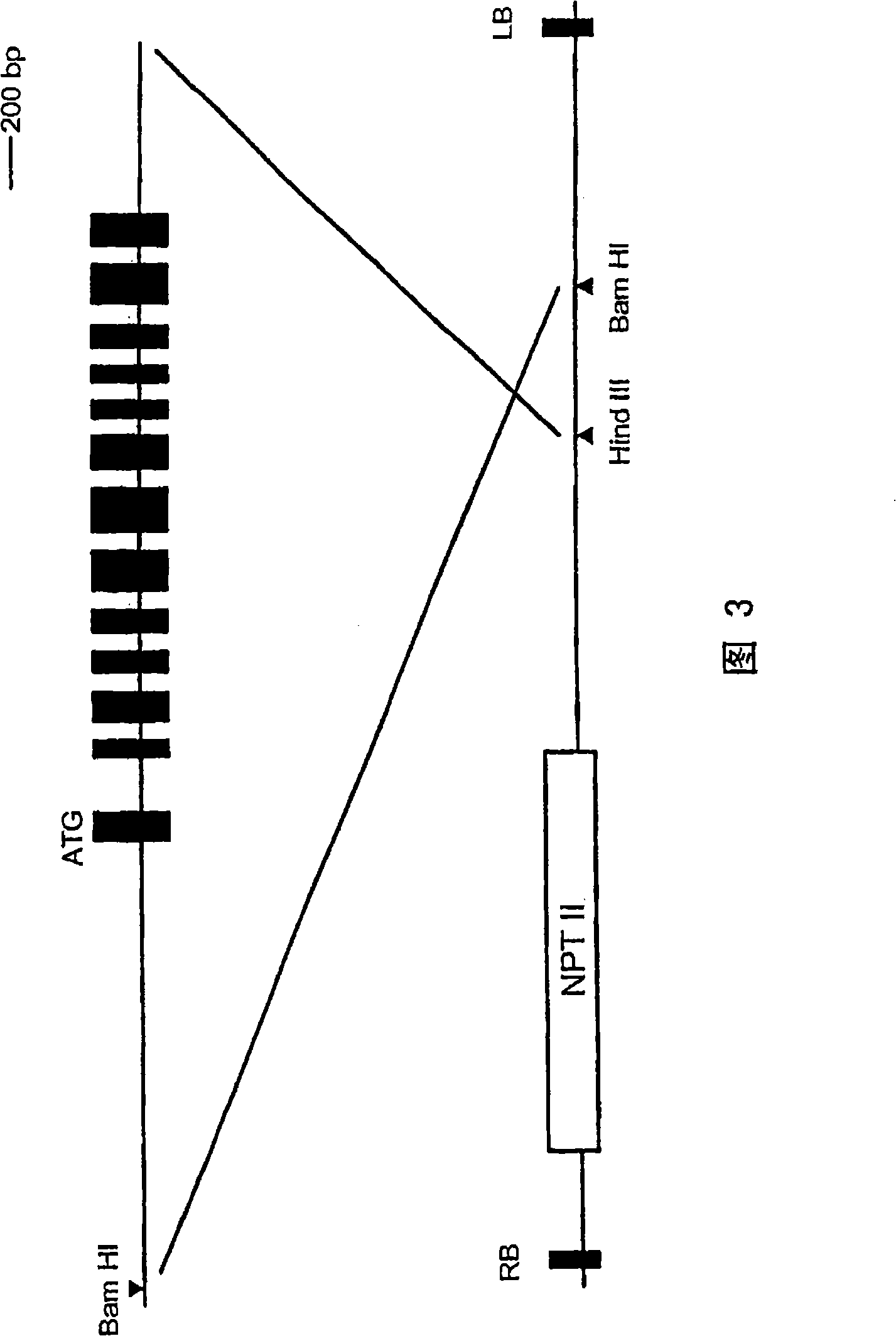
[0088] (1) Preparation of primers for screening GGT deletion strains
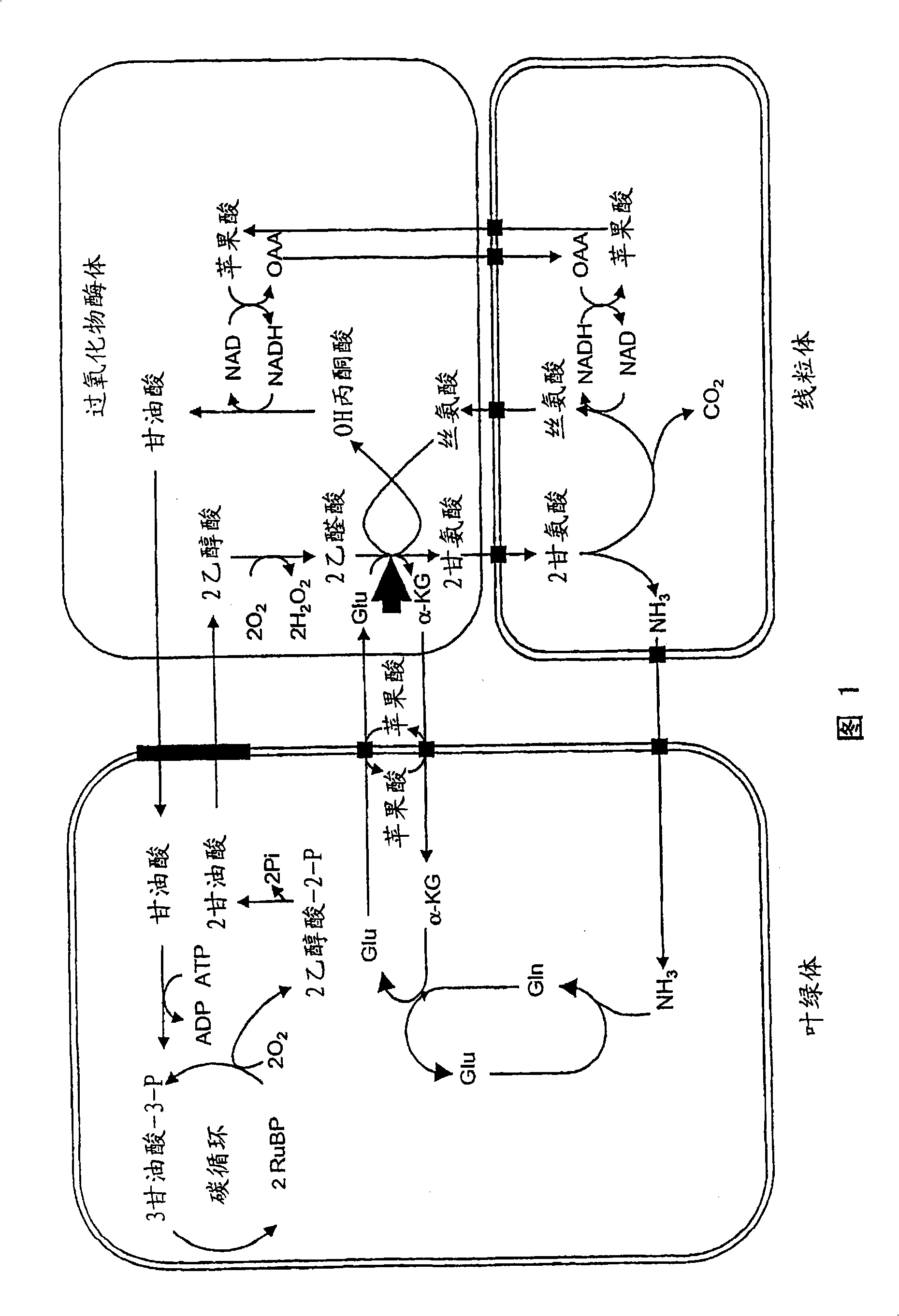
[0089] The GGT gene, since it is also the AlaAT gene, was obtained from the alanine aminotransferase (AlaAT) gene information of Arabidopsis thaliana.
[0090] According to the AlaAT gene data (GenBank) published on the Internet, the copy number and sequence of AlaAT (GGT) were predicted to prepare primers. A data search was performed using alanine aminotransferase and Arabidopsis as keywords, and it was found that there were at least 4 copies of a gene that was predicted to be a GGT gene in the genome. The GenBank accession numbers of each gene are AC005292 (F26F24.16), AC011663 (F5A18.24), AC016529 (T10D10.20), AC026479 (T13M22.3). Each gene is named GGT1, GGT2, GGT3, and GGT4, respectively, and the nucleotide sequences of each cDNA are recorded as sequence numbers 1, 3, 5, and 7, and the predicted amino acid sequence...
reference example 2
[0107] Reference Example 2. Obtaining a GGT Deletion Homozygous Strain
[0108] (1) Screening of homozygotes
[0109] T2 seeds of the line with the confirmed insertion marker were sown on MS medium containing 10 mg / l hygromycin. Transplanted onto asbestos 3 weeks later DNA was extracted from approximately 5 mm square samples of Rossetto leaves. The extraction method used Li's method (Plant J.8: 457-463). To confirm homozygosity, PCR was performed using primers (AAT1U / AAT1L2) sandwiching the marker. PCR was carried out 30 cycles under the conditions of denaturation at 94°C for 30 seconds, annealing at 57°C for 30 seconds, and extension at 72°C for 60 seconds. As a control, wild-type genomic DNA was used as a template. Part of the PCR products were separated by 1% agarose gel electrophoresis. There were 11 homozygous lines among the 35 lines.
[0110] (2) Detection of GGT expression
[0111] The progeny of the obtained homozygous germlines were used to confirm whether g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com