Sperm cell separation methods and compositions containing aptamers or nucleic acid sequences for use therein
一种核酸序列、核苷酸序列的技术,应用在精细胞分离和含有其中所用适体或核酸序列的组合物领域,能够解决费用高精细胞不可逆染色等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
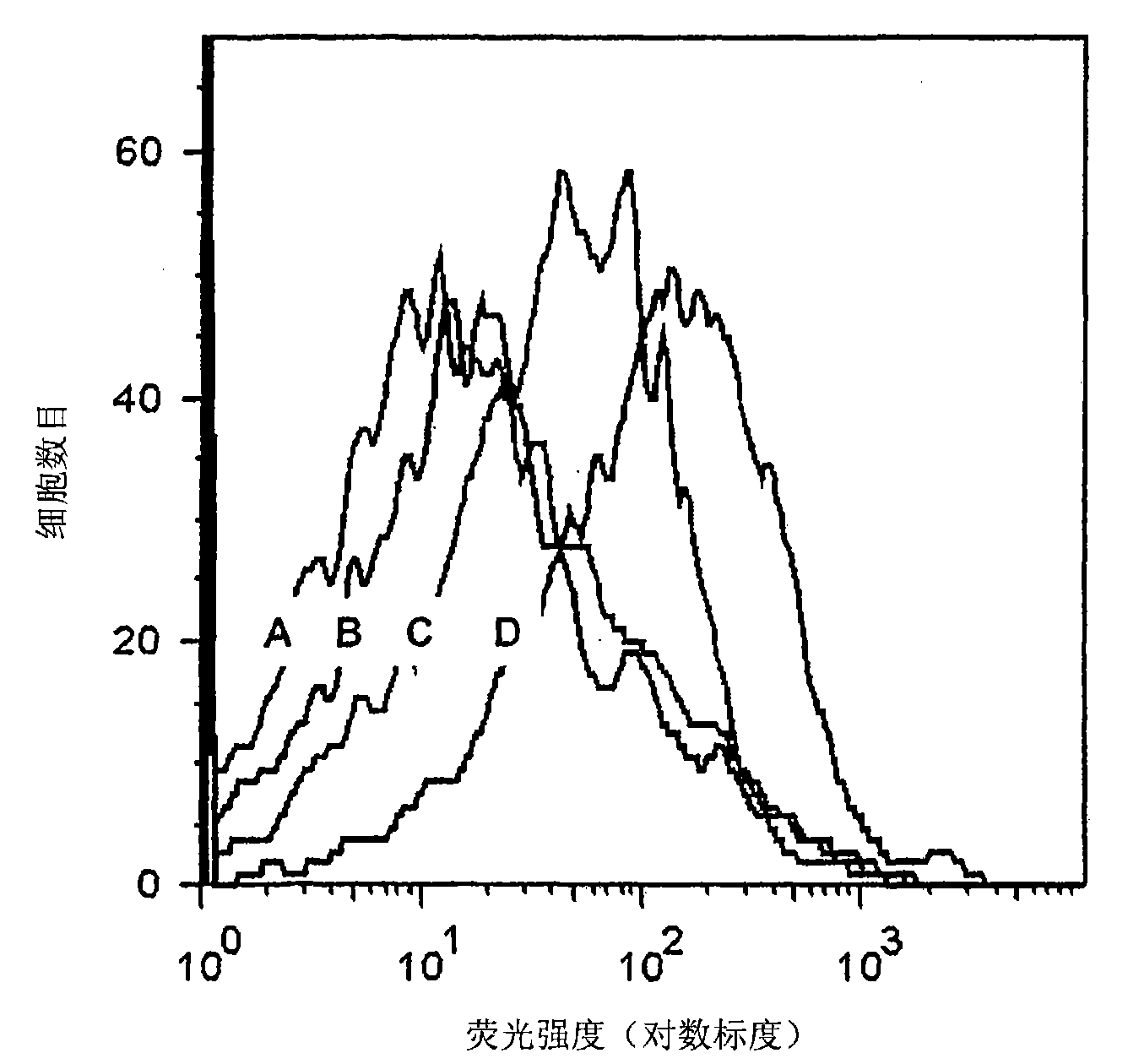
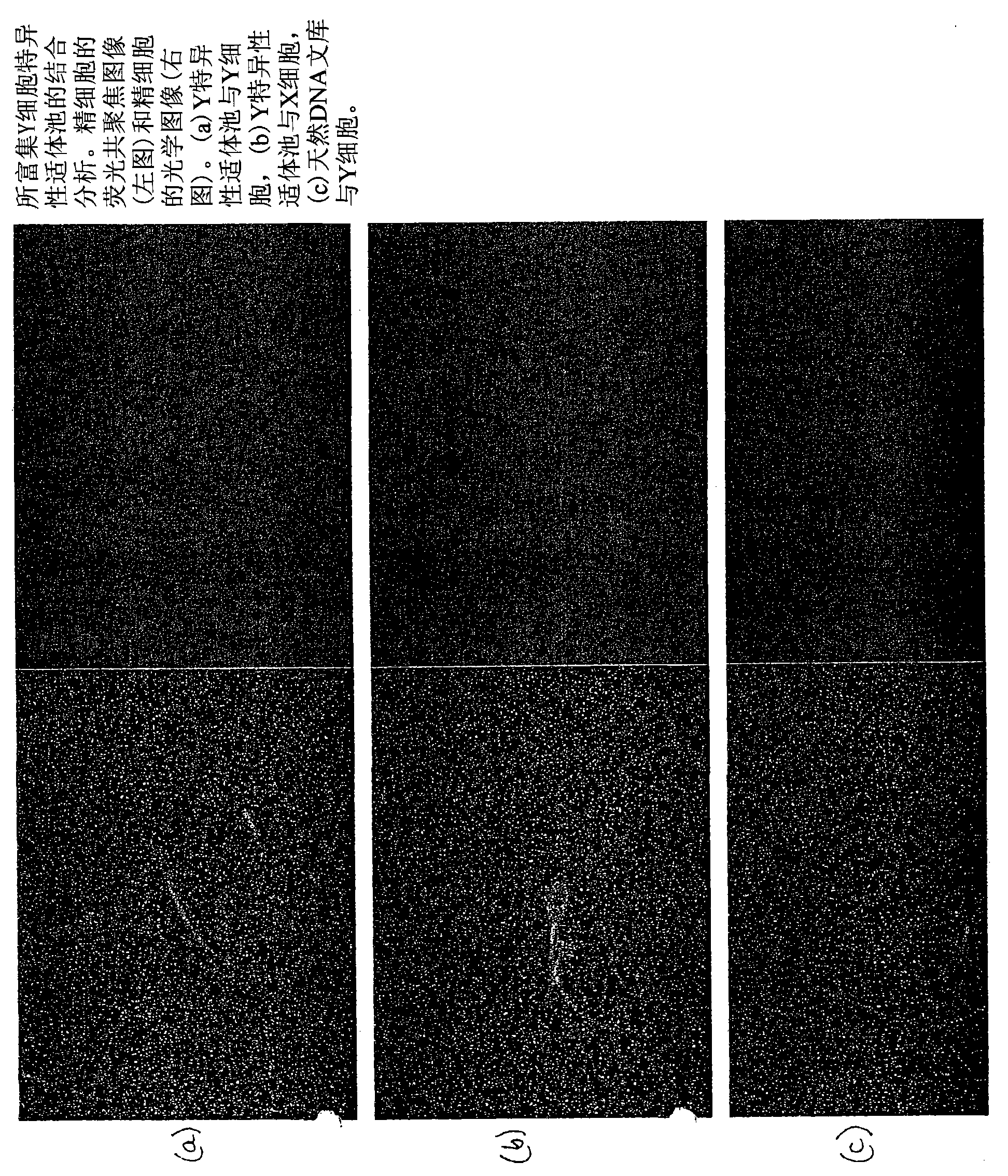
[0062] Sperm aptamer selection method
[0063] The preferred method of selecting specific aptamers of the present invention employs a method that utilizes several rounds of incubation of unsorted, sorted X and / or sorted Y sperm cells with the DNA library and the final step is to collect the solution with the DNA binder and to The solution was used as a template for PCR and strand separation methods on streptavidin beads. Applicants have determined that more than 6 rounds of sperm aptamer selection according to the invention are optimal. Preferably 7, more preferably 8 and even more preferably 9 rounds of selection are performed as described below. However, if necessary, more than 9 rounds of selection can be performed to obtain aptamers that bind to specific target molecules. It is understood that the protocols described below can be varied in temperature, volume and other parameters that do not affect the results of the protocol but result in the production of specific apta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com