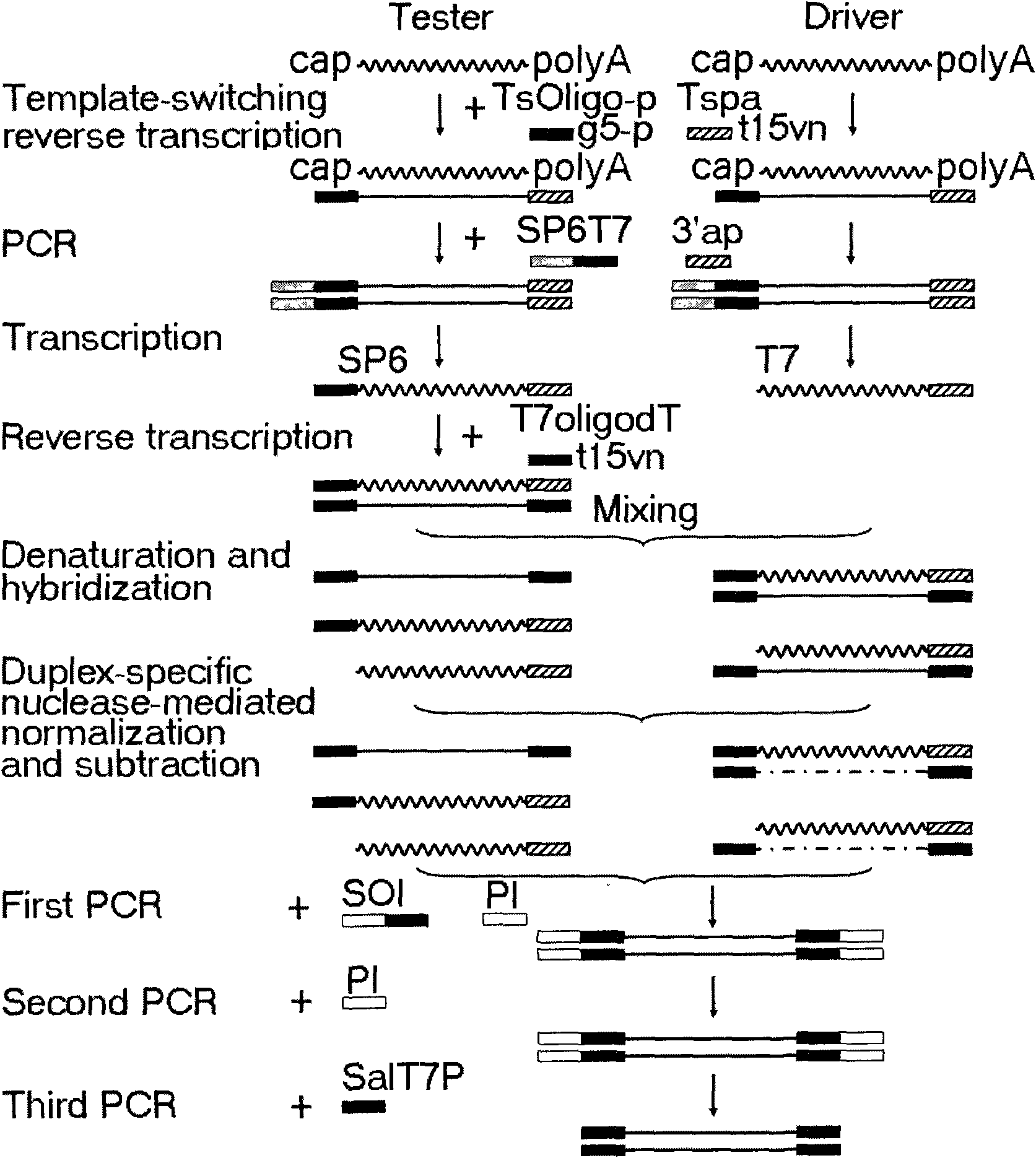
cDNA homogenizing subtractive hybridizing method of double-stranded specific DNA enzyme mediation
A technology of DNase and hybridization method, which is applied in the field of cDNA homogenization subtraction hybridization, can solve the problems that non-target molecules cannot be subtracted, information is easily lost, and SSH is easy to lose information of genes with low expression levels, etc., and achieves high subtraction efficiency. , Efficient enrichment effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Analysis of Artemia dormant reproduction-specific expression genes
[0036] The sequences of the primers (10 μM) used are shown in Table 1:
[0037] Primer
Sequence (5'-3')
TsOligo-p
G TAATACGACTCACTATAGGG GG-p
T7oligod T
G TAATACGACTCACTATAGGG GGTTTTTTTTTTTTTTTVN
SOI
CTGCAGCGAACCAATCCCTCTG TAATACGACTCACTATAGGG
P.I.
CTGCAGCGAACCAATCCCTCTG
SalT7P
GATCGTCGACG TAATACGACTCACTATAGGG
SP6T7
CATTTAGGTGACACTATAGAG TAATACGACTCACTATAGGG
Tspa
TGGTTGGACTCGGTTTGGACGCCATAGAATTGGTTTTTTTTTTTTTTTVN
3'ap
TGGTTGGACTCGGTTTGGACG
[0038] Note: The T7 promoter sequence is underlined, and the 3' end of the primer TsOligo-p is phosphorylated.
[0039] The purpose of this experiment is to isolate genes specifically expressed in dormant reproduction in Artemia parthenogenetica (Gahai Lake, China).
[0040] (1) Synthesizing the first-strand cDNA.
[0041] Two groups of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com